Sanju Sinha, Assistant Professor at Sanford Burnham Prebys, shared a post on X:
“Cancer research currently lacks the concept of ‘timing a molecular event’.
How long ago did this mutation occur?
Years, months or decades?
There’s a clever trick to do this though (partially).
While cancer research have a way to tell order of molecular event in a tumor (early vs. late), we mostly lack a way to say how long ago it happened.
In other words, we miss a Radioactive dating for a molecular alterations.
Why should you even care?
I argue this is very important. If one can tell when the first ‘driver mutation’ occurred, we can date at what age pre-cancer lesion started.
This info can shape our Efforts towards Screening, preventive interventions, and understanding cancer risks.
This question has been bothering me since years (Note from my ideas doc from years ago);
A discussion with Samprita Das revealed that there’s theoretically way to do this.
+A paper already showed the proof of concept – the results are so cool.

In 2020, the following paper came out.
While not too exciting looking at first look, this paper presented an ingenious section called – ‘Chronological time estimates’ to time how long ago a molecular change happened.
How?
Just like tree rings tell time in trees, They noted that cells have natural clocks – CpG-to-TpG mutations – the most common mutation in normal or cancer cells.
See this mutation signature (SBS1) vs. age across many tumor types.
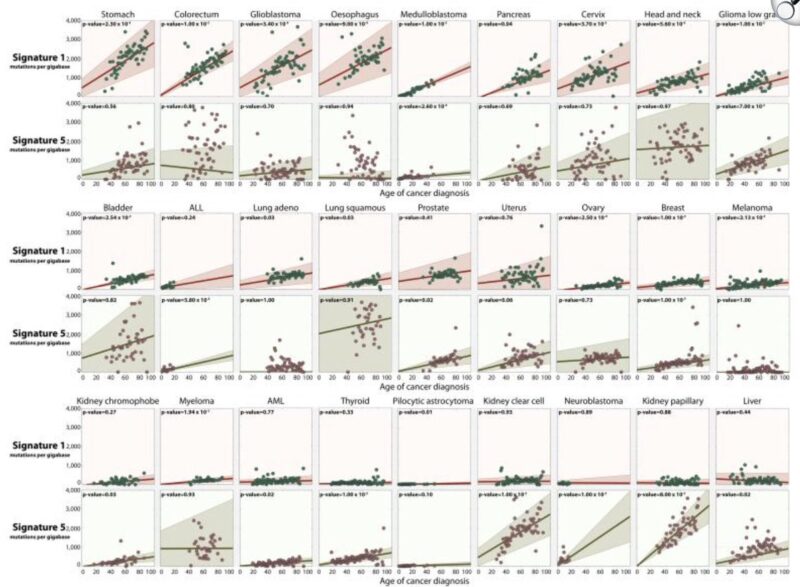
We us this molecular clock to time big amplifications events? eg. whole genome doubling.
When cancer duplicates its genome, mutations present before duplication appear on all copies, while later mutations don’t. By comparing these ratios, we can reconstruct WHEN events happened.
But there’s an issue – cancer cells can still speed up their mutation clock. Emperical data shows that this can be as high as 2.5-7.5x faster.
Jury is still not out on this. Need more data.
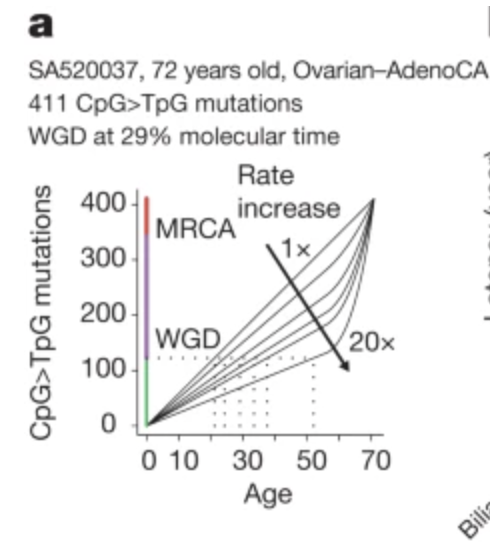
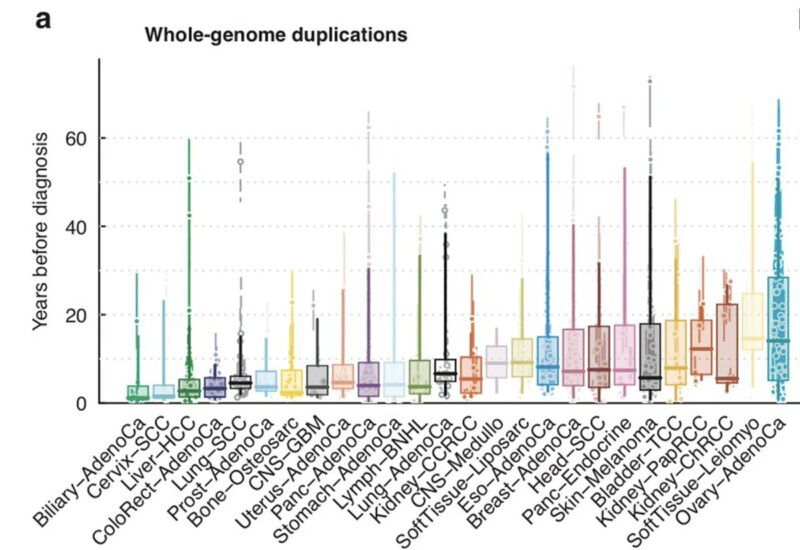
Modeling this range, we can still estimate the time of first genome-duplications and i absolutely love this plot.
Just look at this – Y-axis is estimated years when genome-duplication occurred before clinical cancer diagnosis.
Look at ovary.
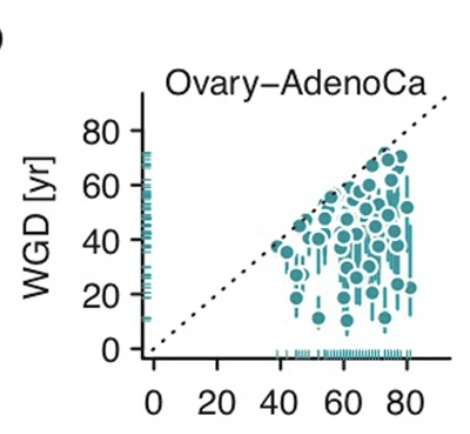
Just look at this data – The time of diagnosis (x axis) and inferred time of genome-duplication (y axis).
Cells acquiring duplication as early as when one is 10-12 years old may have been cancer cell of origin.
One can use this to infer real-time estimate of other events as well;
This is yet low-confidence in my opinion but shows what’s possible.
This analysis can estiamte/teach us how long ‘lesions’ stay dormant before clinical symptoms.
Look at the range of y-axis. Lesions of multiple cancer types might be latent for more a decade.
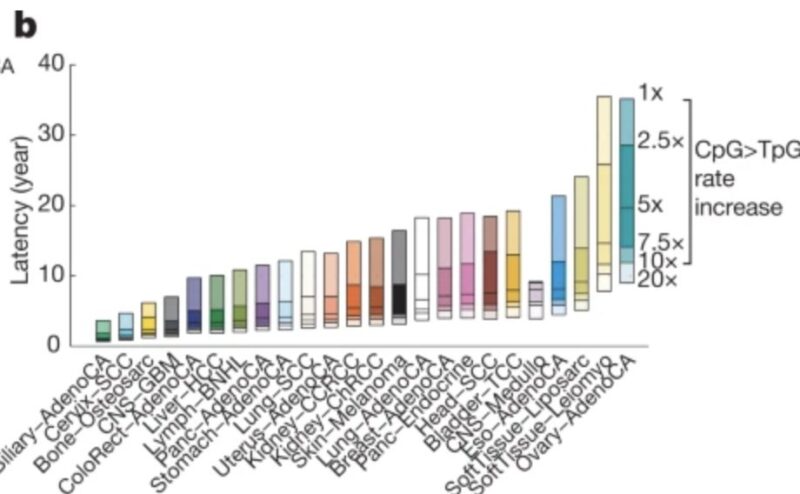
These age number might yet be underestimates, as bulk only captures mutations with large-number of cells (last few generations with few cells are not counted).
So what do we learn?
First, this just starts the problems – We have yet not resolved how to precisely say “when” an event has occurred aside of genome-dupliations. (Q1)
Second, if this is correct, what other precise pieces are needed to trigger these ‘lesions’? (Q2)
Ending with an appreciation post of Ovary: It’s a remarkably organized tissue dynamically re-modelling itself every cycle. eg. >1K follicles at different developmental stages coexist + multiple corpora lutea – all in same physical space.
(needs a diff thread – prob tomorrow).”


