
Eduard Reznik: Selection of cancer driver mutations influenced by obesity
Eduard Reznik, Assistant Attending Computational Oncologist at Memorial Sloan Kettering Cancer Center, shared a post on X:
“So happy to report a new discovery relating obesity to the selective pressure for driver mutations, out now in Nature Genetics.
This is the latest science from all-star Cerise Tang with help from soon-to-be all-star Venise Jan Castillon, and co-led by Justin Jee.
Obesity-dependent selection of driver mutations in cancer
Authors: Cerise Tang, Venise Jan Castillon, Michele Waters, Chris Fong, Tricia Park, Sonia Boscenco, Susie Kim, Kelly Pekala, Jian Carrot-Zhang, A. Ari Hakimi, Nikolaus Schultz, Irina Ostrovnaya, Alexander Gusev, Justin Jee and Ed Reznik.

Obesity is among the leading risk factors for the development of cancer + of prognostic/therapeutic value. We reasoned the obese state may predispose patients to present with molecularly distinct tumors. Reasonable hypothesis, unexpectedly impossible to test. Google it yourself.
There are no existing large-scale cohorts capturing weight/BMI + genomic data. Enter Dr. Jee + his incredible team from Memorial Sloan Kettering Cancer Center‘s Cancer Data Science Initiative, who went to the clinical record of MSK’s sequenced patients and extracted BMI information.
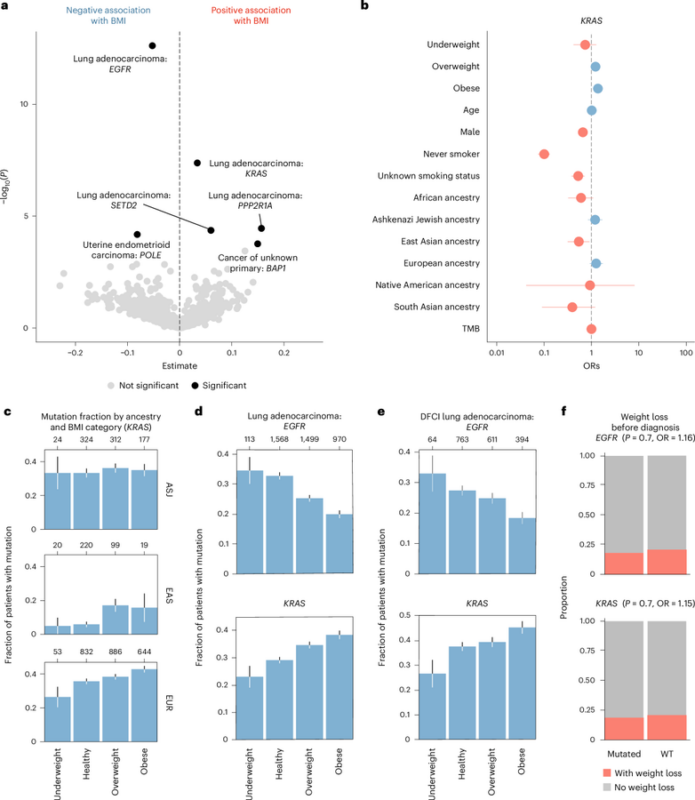
We discovered that certain genomic alterations, most prominently in lung cancer, are associated with BMI at time of sequencing. EGFR elevated at low BMI, KRAS at high BMI. This was effect independent of typical clinical covariates like age and sex.
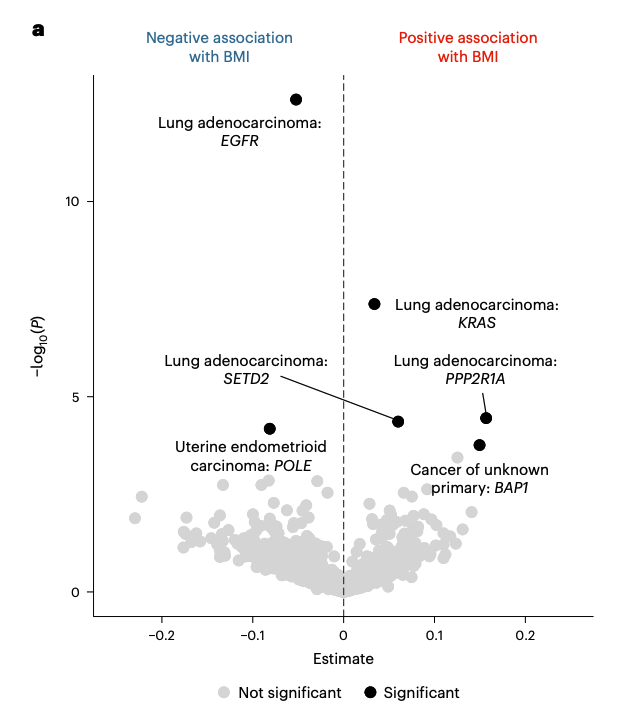
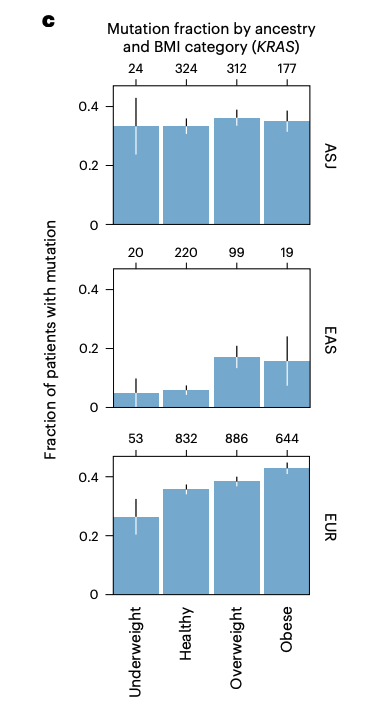
Driver mutations are known to accumulate at different rates in patients of distinct genetic ancestries, raising the possibility that an association between tumor genotypes and obesity status could emerge indirectly. Nope. Also, I love error bars.
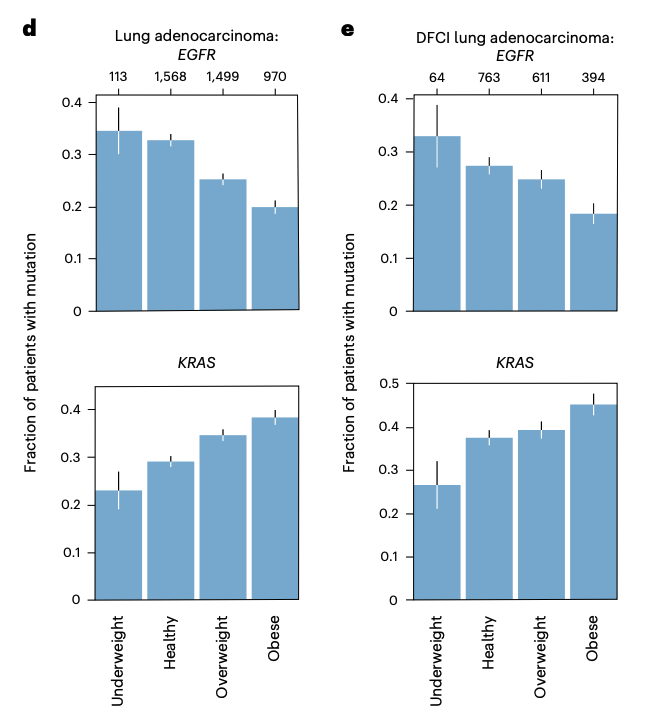
I shamelessly cornered Sasha Gusev at a conference to see if he could help corroborate our findings with DFCI’s data. He emailed me 6 hours later with the results. Need I say more? Also still loving the error bars.
The reviewers kept it spicy and asked us to control for air pollution exposure and socioeconomic status. So we did, but our results remained robust and significant. This may have been the moment where I said wow out loud.
Finally, we ruled out the possibility this is caused by cachexia. More on the relationship between cachexia and cancer genotype to come, look out Pershing Square Sohn Cancer Research Alliance.
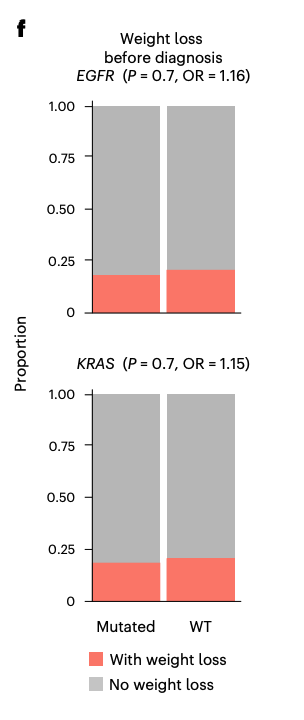
You may be asking yourself so what? Thank you for doing so. A few so what’s here.
These findings suggest that in certain diseases, especially lung cancer, obesity modifies cancer risk in a way that sculpts the dominant molecular subtype of disease.
How?
We don’t know yet. One possibility among many is that it modifies the selective advantage of pre-existing driver alleles in preneoplasia. How? Altered immunosurveillance, altered metabolism, or something else. We aren’t the first to posit this, see.
Organismal metabolism regulates the expansion of oncogenic PIK3CA mutant clones in normal esophagus
Authors: Albert Herms, Bartomeu Colom, Gabriel Piedrafita, Argyro Kalogeropoulou, Ujjwal Banerjee, Charlotte King, Emilie Abby, Kasumi Murai, Irene Caseda, David Fernandez-Antoran, Swee Hoe Ong, Michael W. J. Hall, Christopher Bryant, Roshan K. Sood, Joanna C. Fowler, Albert Pol, Christian Frezza, Bart Vanhaesebroeck and Philip H. Jones.
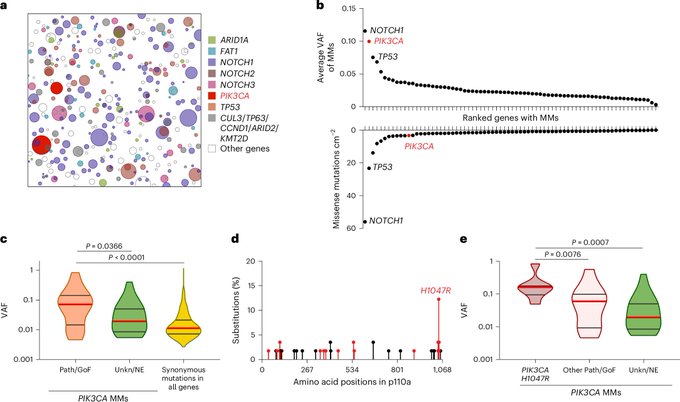
I will add that this hypothesis is particularly alluring given the GLP-1/cancer risk vibes going around these days.
Not sure the data is there to test this yet, but it may be soon.
A final thank you as always to the patients, without whom we would not be able to carry out this scale of science.
We’re excited now to see what comes next.”
-
Challenging the Status Quo in Colorectal Cancer 2024
December 6-8, 2024
-
ESMO 2024 Congress
September 13-17, 2024
-
ASCO Annual Meeting
May 30 - June 4, 2024
-
Yvonne Award 2024
May 31, 2024
-
OncoThon 2024, Online
Feb. 15, 2024
-
Global Summit on War & Cancer 2023, Online
Dec. 14-16, 2023
