
10 Top Translational Papers in GU Oncology suggested by Toni Choueiri
Toni Choueiri, Director of the Lank Center for GU Oncology at Dana-Farber Cancer Institute, shared 10 articles on X:
“It is this time of the Year again: 10 Top Translational Papers in GU Oncology.
Prostate Paper 1:
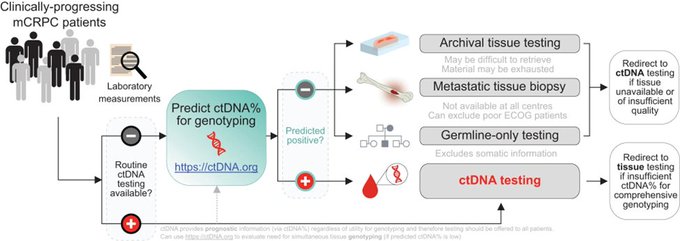
Fonseca et al. introduced a machine-learning approach that predicts whether patients with metastatic castration-resistant prostate cancer (mCRPC) have sufficient circulating tumor DNA fraction (ctDNA%) for informative genotyping.
This study addresses a critical clinical need given that ctDNA-based biomarker genotyping can be limited by low ctDNA in Nature Communications.
Authors: Nicolette M. Fonseca, Corinne Maurice-Dror, Cameron Herberts, Wilson Tu, William Fan, Andrew J. Murtha, Catarina Kollmannsberger, Edmond M. Kwan, Karan Parekh, Elena Schönlau, Cecily Q. Bernales, Gráinne Donnellan, Sarah W. S. Ng, Takayuki Sumiyoshi, Joanna Vergidis, Krista Noonan, Daygen L. Finch, Muhammad Zulfiqar, Stacy Miller, Sunil Parimi, Jean-Michel Lavoie, Edward Hardy, Maryam Soleimani, Lucia Nappi, Bernhard J. Eigl, Christian Kollmannsberger, Sinja Taavitsainen, Matti Nykter, Sofie H. Tolmeijer, Emmy Boerrigter, Niven Mehra, Nielka P. van Erp, Bram De Laere, Johan Lindberg, Henrik Grönberg, Daniel J. Khalaf, Matti Annala, Kim N. Chi and Alexander W. Wyatt
Prostate Paper 2:
Franseschini et al. developed a targeted DNA methylation assay for the detection of neuroendocrine prostate cancer using plasma cell-free DNA with an AUC > 0.93.
Their assay also quantifies tumor content and offers a phenotype evidence score, which reflects various CRPC phenotypes in Cancer Discovery.
Title: Noninvasive Detection of Neuroendocrine Prostate Cancer through Targeted Cell-free DNA Methylation
Authors: Gian Marco Franceschini, Orsetta Quaini, Kei Mizuno, Francesco Orlando, Yari Ciani, Sheng-Yu Ku, Michael Sigouros, Emily Rothmann, Alicia Alonso, Matteo Benelli, Caterina Nardella, Joonghoon Auh, Dory Freeman, Brian Hanratty, Mohamed Adil, Olivier Elemento, Scott T. Tagawa, Felix Y. Feng, Orazio Caffo, Consuelo Buttigliero, Umberto Basso, Peter S. Nelson, Eva Corey, Michael C. Haffner, Gerhardt Attard, Ana Aparicio, Francesca Demichelis and Himisha Beltran

Prostate Paper 3
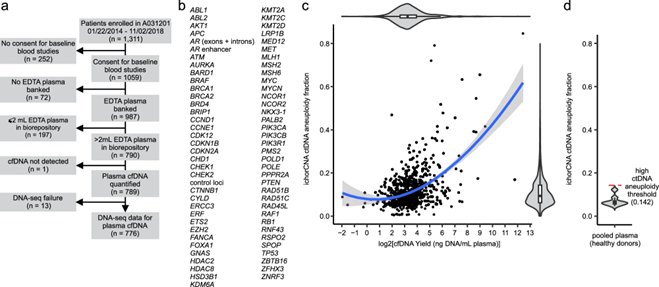
Nutson et al. developed the AR-ctDETECT assay to detect ctDNA in limiting plasma cfDNA from patients with mCRPC in Alliance for Clinical Trials in Oncology A031201 phase 3 trial of enzalutamide with or without abiraterone.
The ctDNA assay was able to prognosticate outcomes, as patients with mCRPC who were ctDNA-positive vs negative had significantly worse median OS (29.0 vs. 47.4 months, respectively) in Nature Communications.
Authors: Todd P. Knutson, Bin Luo, Anna Kobilka, Jacqueline Lyman, Siyuan Guo, Sarah A. Munro, Yingming Li, Rakesh Heer, Luke Gaughan, Michael J. Morris, Himisha Beltran, Charles J. Ryan, Emmanuel S. Antonarakis, Andrew J. Armstrong, Susan Halabi and Scott M. Dehm

Kidney Paper 4:
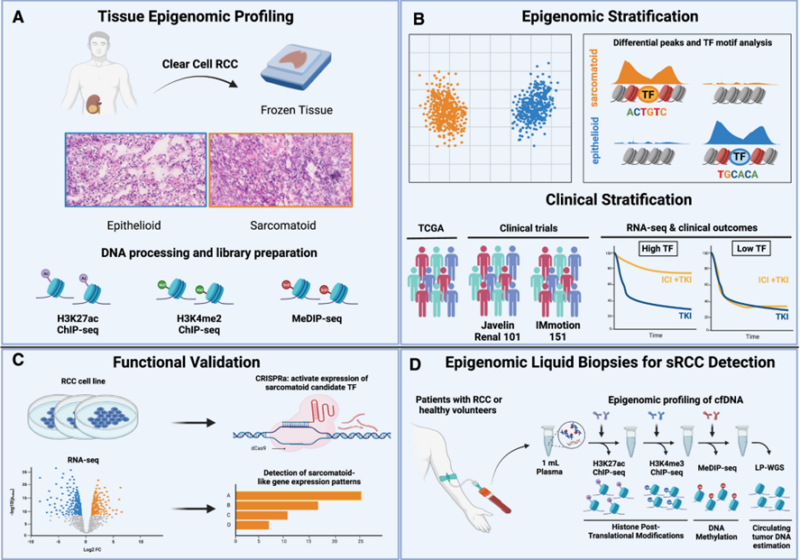
Our very own Talal El Zarif and Karl Semaan described the epigenetic landscape of sarcomatoid RCC, identifying transcriptional programs associated with patient outcomes and response to immunotherapy.
They have also established a blood-based diagnostic approach using detectable sarc-RCC epigenomic signatures in patient plasma.
Title: Epigenomic signatures of sarcomatoid differentiation to guide the treatment of renal cell carcinoma
Authors: Talal El Zarif1, Karl Semaan, Marc Eid, Ji-Heui Seo, Simon Garinet, Matthew P. Davidsohn, Pranshu Sahgal, Brad Fortunato, John Canniff, Amin H. Nassar, Sarah Abou Alaiwi, Ziad Bakouny, Gitanjali Lakshminarayanan, Hunter Savignano, Kevin Lyons, Sayed Matar, Atef Ali, Eddy Saad, Renee Maria Saliby, Paulo Cordeiro, Ziwei Zhang, Nourhan El Ahmar, Yasmin Nabil Laimon, Chris Labaki, Valisha Shah, Dory Freeman, Jillian O’Toole, Gwo-Shu Mary Lee, Justin Hwang, Mark Pomerantz, Sabina Signoretti, Eliezer M. Van Allen, Wanling Xie, Jacob E. Berchuck, Srinivas R. Viswanathan, David A. Braun, Toni K. Choueiri, Matthew L. Freedman and Sylvan C. Baca

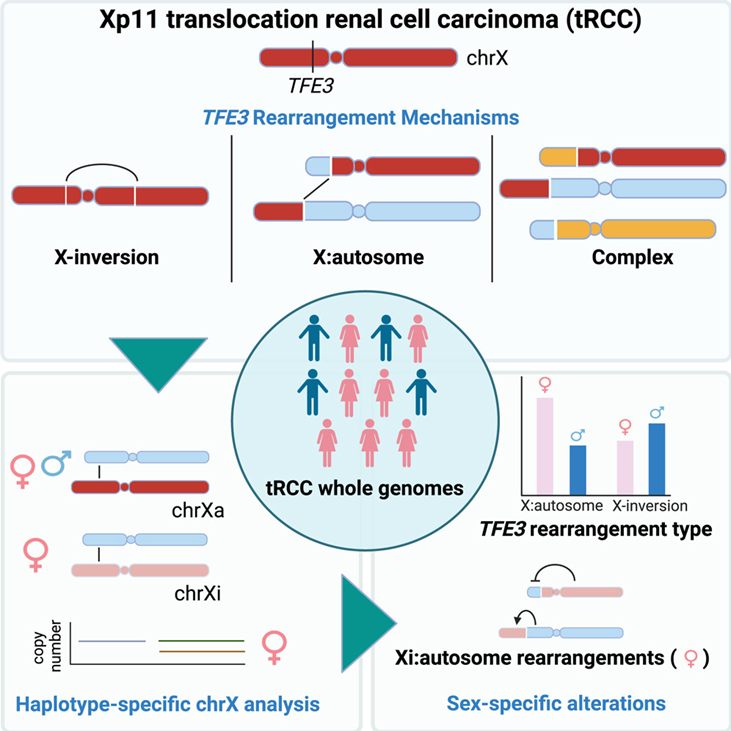
Kidney Paper 5:
Achom et al. conducted haplotype-specific analyses of chromosome X rearrangements in tRCC whole genomes, demonstrating that TFE3 fusions consistently occur as reciprocal translocations. They also demonstrated that oncogenic TFE3 fusions can originate from chromosome Xi:autosomal translocations and that female-specific chrXi:autosomal translocations lead to a 2:1 female-to-male ratio of TFE3 fusions with autosomal partner genes, explaining the female predominance in tRCC.
Title: A genetic basis for sex differences in Xp11 translocation renal cell carcinoma
Authors: Mingkee Achom, Ananthan Sadagopan, Chunyang Bao, Fiona McBride, Jiao Li, Prathyusha Konda, Richard W. Tourdot, Qingru Xu, Maria Nakhoul, Daniel S. Gallant, Usman Ali Ahmed, Jillian O’Toole, Dory Freeman, Gwo-Shu Mary Lee, Jonathan L. Hecht, Eric C. Kauffman, David J. Einstein, Toni K. Choueiri, Cheng-Zhong Zhang and Srinivas R. Viswanathan

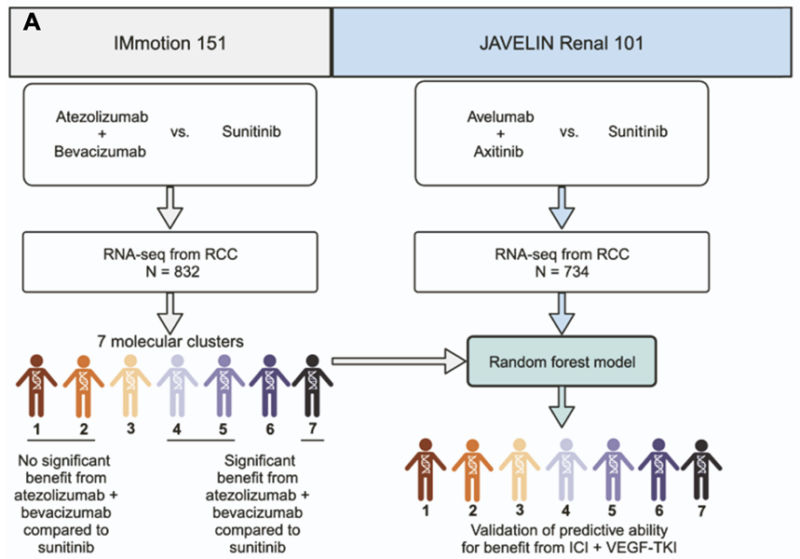
Paper 6 Kidney:
The paper in Cancer Cell by our very own Renee Maria Saliby et al (now at Yale Internal Medicine) showed that distinct molecular RCC subtypes can be accurately classified by a machine-learning model using transcriptomic data.
The model was trained on the RNA-seq data from the IMmotion 151 trial and then applied to classify 734 tumors from the JR101 trial into one of seven molecular subtypes.
Title: Impact of renal cell carcinoma molecular subtypes on immunotherapy and targeted therapy outcomes
Authors: Renée Maria Saliby, Chris Labaki1, Tejas R. Jammihal, Wanling Xie, Maxine Sun, Valisha Shah, Eddy Saad, M. Harry Kane, Soki Kashima, Katherine Sadak, Talal El Zarif, Deepak Poduval, Robert J. Motzer, Thomas Powles, Brian I. Rini, Laurence Albiges, Sumanta K. Pal, Bradley A. McGregor, Rana R. McKay, Sabina Signoretti, Eliezer M. Van Allen, Sachet A. Shukla, Toni K. Choueiri and David A. Braun

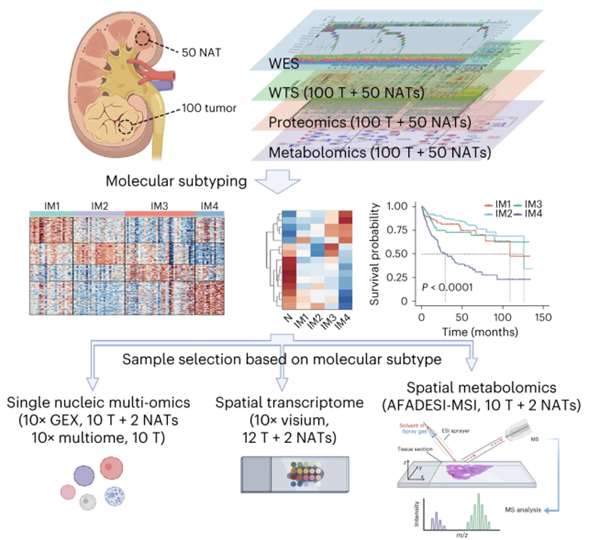
Paper 7 Kidney:
Junyi Hu et al. published a comprehensive, multi-omic profiling analysis including genomic, transcriptomic, proteomic, spatial transcriptomic and metabolomic analyses of 100 clear-cell RCC patients in Nature Genetics, introducing four different ccRCC subtypes and suggesting a treatment strategy based on subtype-specific immune cell infiltration.
Authors: Junyi Hu, Shao-Gang Wang, Yaxin Hou, Zhaohui Chen, Lilong Liu, Ruizhi Li, Nisha Li, Lijie Zhou, Yu Yang, Liping Wang, Liang Wang, Xiong Yang, Yichen Lei, Changqi Deng, Yang Li, Zhiyao Deng, Yuhong Ding, Yingchun Kuang, Zhipeng Yao, Yang Xun, Fan Li, Heng Li, Jia Hu, Zheng Liu, Tao Wang, Yi Hao, Xuanmao Jiao, Wei Guan, Zhen Tao, Shancheng Ren and Ke Chen

Paper 8 Bladder:
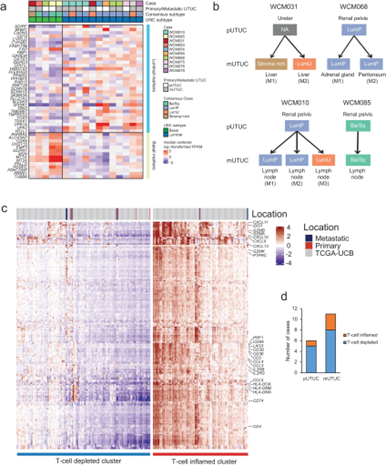
By integrating genomic, transcriptomic, and single-cell protein markers, Ohara et al. show that actionable genomic alterations are often discordant between matched primary and metastatic UTUC tumors, whereas molecular subtypes and signature of immune depletion remain stable.
Work from Bishoy M. Faltas in Nature Communications.
Authors: Kentaro Ohara, André Figueiredo Rendeiro, Bhavneet Bhinder, Kenneth Wha Eng, Hiranmayi Ravichandran, Duy Nguyen, David Pisapia, Aram Vosoughi, Evan Fernandez, Kyrillus S. Shohdy, Jyothi Manohar, Shaham Beg, David Wilkes, Brian D. Robinson, Francesca Khani, Rohan Bareja, Scott T. Tagawa, Madhu M. Ouseph, Andrea Sboner, Olivier Elemento, Bishoy M. Faltas and Juan Miguel Mosquera
Paper 9 Bladder:
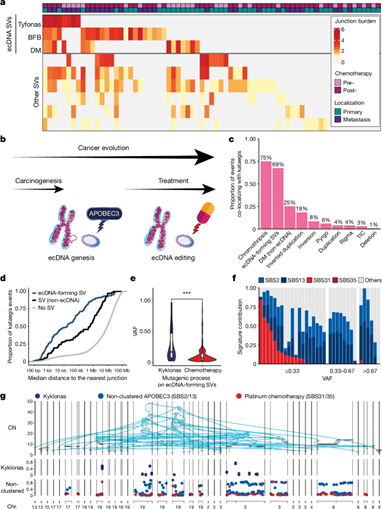
In this outstanding paper in Nature, Nguyen et al. again from Bishoy M. Faltas (what a year for Bishoy M. Faltas) reveal how APOBEC3 drives early clonal mutations in urothelial carcinoma, while chemo induces late subclonal mutations. They identified circular extrachromosomal DNA driving CCND1 amplification and resistance, which evolves by adding DNA segments.
Finally, they map ecDNA architecture and link it to cancer progression. Huge findings that may redefine therapeutic strategies to target ecDNA.
Title: The interplay of mutagenesis and ecDNA shapes urothelial cancer evolution
Authors: Duy D. Nguyen, William F. Hooper, Weisi Liu, Timothy R. Chu, Heather Geiger, Jennifer M. Shelton, Minita Shah, Zoe R. Goldstein, Lara Winterkorn, Adrienne Helland, Michael Sigouros, Jyothi Manohar, Jenna Moyer, Majd Al Assaad, Alissa Semaan, Sandra Cohen, Florencia Madorsky Rowdo, David Wilkes, Mohamed Osman, Rahul R. Singh, Andrea Sboner, Henkel L. Valentine, Phillip Abbosh, Scott T. Tagawa, David M. Nanus, Jones T. Nauseef, Cora N. Sternberg, Ana M. Molina, Douglas Scherr, Giorgio Inghirami, Juan Miguel Mosquera, Olivier Elemento, Nicolas Robine and Bishoy M. Faltas
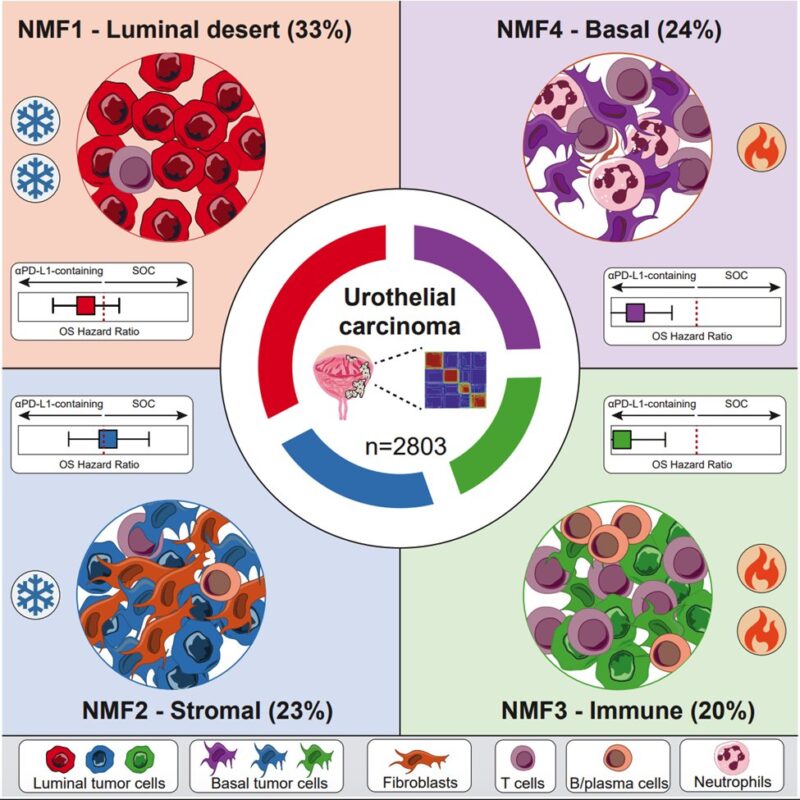
Paper 10 Bladder:
Important work by Hamidi et al. who identified four molecular subtypes in urothelial carcinoma with distinct tumor microenvironments, revealed a survival benefit from atezolizumab for immune and basal tumors, and proposed a digital pathology that predicts molecular subtypes from single H&E slides, potentially tailoring treatment.
Authors: Habib Hamidi, Yasin Senbabaoglu, Niha Beig, Juliette Roels, Cyrus Manuel, Xiangnan Guan, Hartmut Koeppen, Zoe June Assaf, Barzin Y. Nabet, Adrian Waddell, Kobe Yuen, Sophia Maund, Ethan Sokol, Jennifer M. Giltnane, Amber Schedlbauer, Eloisa Fuentes, James D. Cowan, Edward E. Kadel, III, Viraj Degaonkar, Alexander Andreev-Drakhlin, Patrick Williams, Corey Carter, Suyasha Gupta, Elizabeth Steinberg, Yohann Loriot, Joaquim Bellmunt, Petros Grivas, Jonathan Rosenberg, Michiel S. van der Heijden, Matthew D. Galsky, Thomas Powles, Sanjeev Mariathasan and Romain Banchereau
Toni Choueiri is the Director of the Lank Center for Genitourinary (GU) Oncology at Dana-Farber Cancer Institute (DFCI), co-leader of the Kidney Cancer Program at Dana-Farber/Harvard Cancer Center, and the Jerome and Nancy Kohlberg Chair and Professor of Medicine at Harvard Medical School.
As a medical oncologist, clinical trialist, and translational researcher, he specializes in treating genitourinary cancers (prostate, bladder, testis, and kidney cancer), with a focus on kidney cancer.
More posts featuring Toni Choueiri.
-
ESMO 2024 Congress
September 13-17, 2024
-
ASCO Annual Meeting
May 30 - June 4, 2024
-
Yvonne Award 2024
May 31, 2024
-
OncoThon 2024, Online
Feb. 15, 2024
-
Global Summit on War & Cancer 2023, Online
Dec. 14-16, 2023
