Hepatocellular carcinoma (HCC) is the most common primary liver cancer and remains among the top causes of cancer-related deaths worldwide. Although modern systemic therapies such as atezolizumab plus bevacizumab (A+B) and tyrosine kinase inhibitors (TKIs) have improved outcomes, the biological underpinnings of treatment response in advanced disease are still poorly understood.
Because HCC can often be diagnosed radiologically without biopsy, molecular data on its genomic alterations remain limited. This gap has hindered the development of predictive biomarkers and targeted therapies. In an important new analysis published in ESMO Open (Vol. 10, Issue 11, 2025), Salani et al. investigated the genomic landscape of advanced HCC using one of the largest real-world datasets to date, offering valuable insights into how tumor genetics may shape prognosis and treatment response.
Methods
The investigators conducted a retrospective analysis using the Flatiron Health–Foundation Medicine clinico-genomic database, which links electronic health record data from approximately 280 U.S. cancer clinics with comprehensive genomic profiling (CGP) performed by Foundation Medicine.
A total of 370 patients with advanced HCC who received systemic therapy between 2011 and 2024 were included. Both tissue and liquid biopsies were analyzed using next-generation sequencing to identify genomic alterations (GAs) across hundreds of cancer-related genes.
The study assessed the frequency and co-occurrence of mutations, compared viral and non-viral HCC etiologies, and explored their prognostic and predictive associations—particularly in patients treated with A+B or TKIs. Time to progression (TTP) was measured through Kaplan–Meier and Cox proportional hazards analyses, adjusting for clinical variables such as age, sex, performance status, liver function, and etiology.
You can also read about Curative Treatment after Immunotherapy Leads to Excellent Outcomes in Patients with Hepatocellular Carcinoma on OncoDaily.
Results
The results, published in ESMO Open in November 2025, revealed a detailed molecular portrait of advanced hepatocellular carcinoma across the U.S. population. Among 291 tissue and 86 liquid samples analyzed, the most frequently altered genes were the TERT promoter (TERTp, 61.5%), CTNNB1 (34.0%), and TP53 (33.0%), followed by MYC (16.2%) and ARID1A (11.7%). These alterations mainly affected pathways related to cell cycle and apoptosis, DNA damage repair, WNT/β-catenin signaling, and the p53 pathway.
Interestingly, viral-related HCC—often associated with hepatitis B or C—showed enrichment in TERTp, CTNNB1, and WNT pathway alterations, while non-viral HCC linked to alcohol use or metabolic dysfunction more frequently displayed RTK/RAS pathway mutations. This molecular divergence reflects distinct mechanisms of hepatocarcinogenesis between viral and metabolic liver diseases.
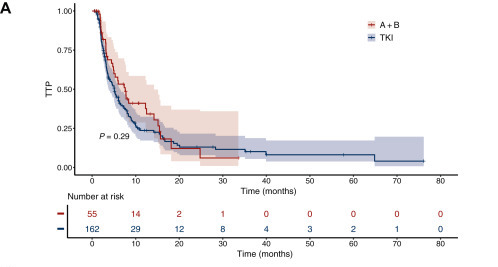
When clinical outcomes were examined, the median TTP for the overall cohort was 5.1 months, with a trend toward longer TTP among patients treated with A+B compared with TKIs (7.6 vs 5.1 months; HR 0.75). Although this difference did not reach statistical significance, it aligned with previous findings from the IMbrave150 trial and other real-world studies.
Certain alterations carried notable clinical implications. TP53 missense mutations—particularly p.V157F and p.R249S—were associated with shorter TTP and poorer prognosis. Conversely, MYC amplification appeared to be a potential negative predictor for A+B therapy, suggesting that MYC-driven tumors might exhibit immune-resistant phenotypescharacterized by low PD-L1 expression and diminished T-cell infiltration. Alterations in chromatin-modifying genes and NOTCH pathway components showed trends toward improved outcomes, whereas DNA damage, p53, and RTK/RASalterations were linked to worse progression dynamics.
Patterns of gene co-occurrence further illuminated HCC biology. Mutations in TERTp often co-existed with CTNNB1and MYC, supporting their synergistic role in hepatocarcinogenesis, while CTNNB1 and RB1 mutations were largely mutually exclusive. Frequent amplifications in FGF3/FGF4/FGF19/CCND1 and MYC loci pointed to converging mechanisms of cell-cycle activation.
Conclusion
This extensive real-world genomic analysis provides one of the most detailed molecular portraits of advanced hepatocellular carcinoma to date. It confirms that TERTp, CTNNB1, and TP53 are the dominant drivers of HCC and that viral and non-viral tumors exhibit distinct genomic fingerprints.
Although most recurrent alterations remain untargetable, their characterization lays the foundation for precision oncology in liver cancer. The potential negative predictive role of MYC in patients receiving atezolizumab plus bevacizumabhighlights the need for deeper exploration of immune-resistant molecular subtypes. Moreover, the frequent involvement of DNA repair and chromatin remodeling pathways suggests new opportunities for investigating PARP inhibitors and synthetic lethality-based strategies in molecularly selected HCC populations.
With comprehensive genomic profiling increasingly accessible, studies like this one bring us closer to identifying the next generation of biomarkers and therapeutic targets that could reshape the future of advanced HCC management.
You can also read about TRIPLET-HCC at ESMO 2025: Adding Ipilimumab to Atezolizumab–Bevacizumab Fails to Improve Outcomes in Unresectable HCC on OncoDaily.