Olivier Elemento, Director of Englander Institute for Precision Medicine at Weill Cornell Medicine, shared a post on LinkedIn:
“AI for Antibody Design: A Tale of Two Architectures
Two exciting preprints dropped recently, showcasing powerful AI-driven methods for generating novel antibodies from scratch. “Germinal” by Luis Santiago Mille Fragoso, Brian Hie, Xiaojing Gao and colleagues, and “mBER” from Erik Swanson, Pierce Ogden and the team at Manifold Bio both cleverly use AlphaFold-Multimer, but their design philosophies and validation strategies offer an interesting comparison.
Germinal’s Approach: A Dynamic Balancing Act
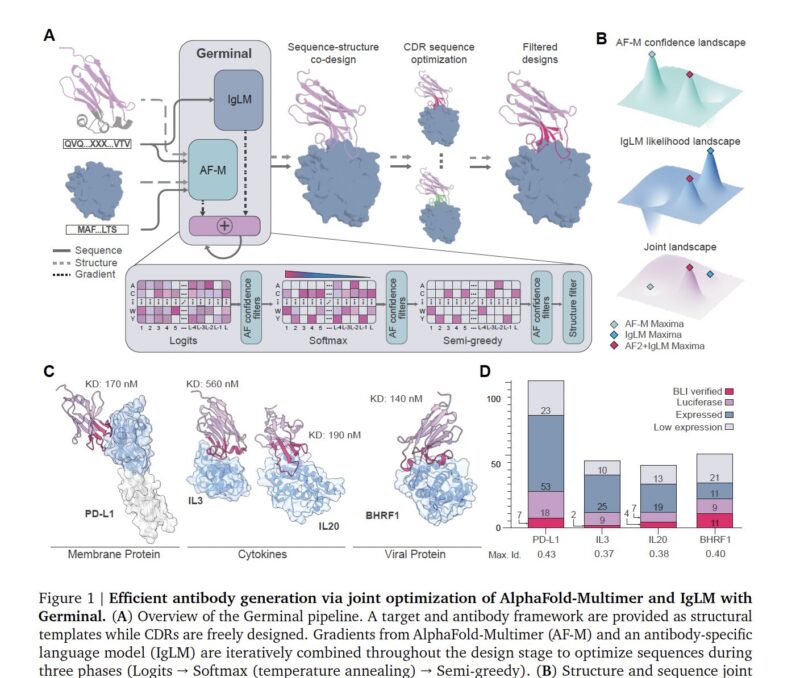
Germinal’s AI acts a bit like a designer trying to satisfy two expert opinions at once. It dynamically balances the goal of structural integrity, using AlphaFold’s rules to ensure a stable 3D shape, with the need for sequence naturalness, using a specialized antibody language model to ensure the sequence looks biologically plausible. Their core insight is that these two goals can often be in conflict. The AI gets continuous feedback from both models, allowing it to navigate the trade-offs and find an optimal design that is both structurally sound and looks like a real antibody.
mBER’s Approach: Designing with a Blueprint
mBER’s strategy is more like giving the AI a detailed instruction manual before it starts. It provides two key pieces of information upfront: a sequence “cheat sheet” of plausible amino acids for the binding regions (generated by the ESM2 model) and a 3D structural scaffold that serves as a blueprint for the antibody’s stable framework. This effectively tells the AI what materials to use and what the final product should generally look like. I think this is a very practical and scalable approach, showing that you can create format-specific antibodies by giving existing AI models the right starting constraints.
Validation: Precision vs. Massive Scale
The divergence in their validation methods is just as interesting. Germinal opted for a precise, in-depth approach. They tested a small number of designs (43-101 per target) for four antigens, using a rapid screening assay to identify promising candidates which were then confirmed with bio-layer interferometry (BLI) to have high-affinity (nanomolar) binding. This resulted in an impressive 4-22% success rate.
In contrast, mBER went for massive scale. They designed over 1 million VHH binders and screened them against 145 targets using a high-throughput phage display platform. Their results showed success against 45% of the tested targets, with success rates for the best-designed sites reaching as high as 38% after filtering designs with a computational confidence score.
Both papers powerfully demonstrate that AI-driven de novo antibody design is rapidly maturing, opening incredible possibilities for therapeutics and research.
More posts featuring Olivier Elemento on OncoDaily.


