At the ESMO Congress 2025 in Berlin, Dr. Martina Carullo (Pisa, Italy) presented groundbreaking findings from the translational program of AtezoTRIBE and AVETRIC, two clinical trials exploring immunotherapy in proficient mismatch repair (pMMR) metastatic colorectal cancer (mCRC). Using the Lunit SCOPE IO artificial intelligence (AI) platform, the investigators developed and validated a digital biomarker capable of predicting which pMMR tumors benefit from immune checkpoint inhibition—an area long considered resistant to immunotherapy.
Background
Immune checkpoint inhibitors (ICIs) have transformed treatment outcomes for patients with deficient mismatch repair (dMMR/MSI-H) mCRC, but have shown minimal activity in pMMR tumors, which constitute the vast majority of metastatic cases. Identifying predictive markers within this refractory group remains one of the central challenges in gastrointestinal oncology.
Recent advances in AI-driven pathology have enabled quantitative analysis of the tumor microenvironment on digitized hematoxylin and eosin (H&E) slides. By mapping immune, stromal, and tumor cell populations, AI models can capture subtle biological patterns invisible to traditional pathology. The present study used this approach to generate an AI-derived biomarker that could distinguish responders from non-responders to ICI-based regimens in pMMR mCRC.
Methods
The analysis incorporated pre-treatment tumor slides from patients enrolled in AtezoTRIBE (NCT03721653) and AVETRIC (NCT04513951). In AtezoTRIBE, patients received FOLFOXIRI/bevacizumab with or without atezolizumab, while in AVETRIC, the regimen was FOLFOXIRI/cetuximab/avelumab.
Using the Lunit SCOPE IO platform, the research team quantified the density of lymphocytes, fibroblasts, macrophages, endothelial, mitotic, and tumor cells within both cancer areas and surrounding stroma. A multivariate Cox regression model was trained on the atezolizumab-treated arm of AtezoTRIBE to identify cellular features most predictive of progression-free survival (PFS). A cut-off optimized for PFS was then applied to classify tumors as biomarker-high or biomarker-low, with AVETRIC serving as an external validation set.
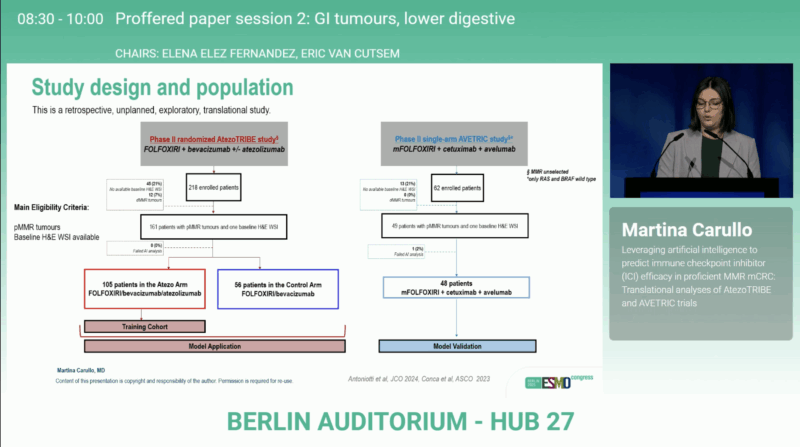
Results from AtezoTRIBE
The AI-based analysis was conducted on whole-slide images from 161 patients. The resulting biomarker integrated densities of tumor and mitotic cells in the cancer area, lymphocytes in the tumor core, and fibroblasts, macrophages, and endothelial cells in the stroma. Of the evaluated patients, 113 (70%) were classified as biomarker-high, a group characterized by older age (p = 0.030) and a higher frequency of liver metastases (p = 0.023).
In the atezolizumab arm, biomarker-high patients achieved significantly superior outcomes compared with biomarker-low ones, with PFS p = 0.036 and overall survival (OS) p = 0.024. No such association was observed in the control arm (PFS p = 0.564; OS p = 0.186).
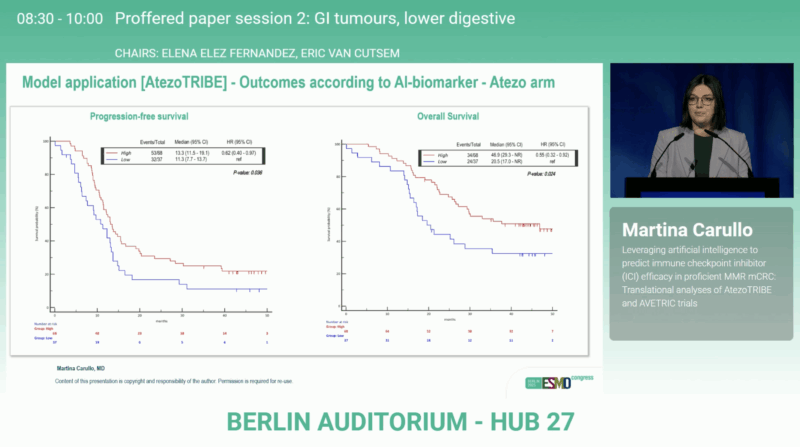
A formal treatment–biomarker interaction analysis showed a stronger benefit from atezolizumab among biomarker-high patients (HR for PFS 0.69; 95% CI 0.45–1.04 and HR for OS 0.54; 95% CI 0.33–0.88), while biomarker-low tumors did not derive advantage (HR for PFS 1.34; 95% CI 0.66–2.72 and HR for OS 1.70; 95% CI 0.69–4.20).
Validation in AVETRIC
The model was independently tested on 48 patients from the AVETRIC trial. Thirty-six (75%) were classified as biomarker-high and displayed numerically improved outcomes compared with biomarker-low cases, with PFS p = 0.043and OS p = 0.053. The validation confirmed the reproducibility of the AI-generated signature across different ICI-based regimens and patient populations.
Interpretation
This dual-trial analysis demonstrates that an AI-defined histologic signature reflecting immune–stromal interactions can predict the benefit of immunotherapy even in microsatellite-stable disease. The biomarker captures a complex interplay between immune infiltration and stromal architecture—features often underestimated by genomic profiling alone.
By transforming standard H&E slides into predictive, quantifiable datasets, this work illustrates how digital pathology and AI can refine patient selection for immunotherapy and accelerate the transition toward precision oncology in pMMR mCRC.
You can read the full abstract here.
Conclusion
The AtezoTRIBE and AVETRIC translational analyses show that AI-derived tumor microenvironment biomarkerscan identify subsets of pMMR colorectal cancer patients who benefit from ICI-based therapy. In AtezoTRIBE, biomarker-high status correlated with significantly longer PFS (p = 0.036) and OS (p = 0.024) under atezolizumab, and these findings were validated in AVETRIC (PFS p = 0.043; OS p = 0.053).

These results mark an important advance in AI-assisted immuno-oncology, suggesting that digital pathology could soon complement molecular testing in guiding treatment for colorectal cancer beyond MSI status.


