Turajlic Lab shared a post on X:
“How do the tumor genome, transcriptome, and TME jointly evolve in clear cell renal cell carcinoma (ccRCC)?
As part of TRACERx Renal, we show in our latest Cancer Discovery study underappreciated non-genetic intratumor heterogeneity in ccRCC.
Here’s what we found.
Non-genetic evolution plays a pivotal role in cancer progression.
Yet, in ccRCC, while genetic evolution is well-explored, how phenotypes and the tumor microenvironment (TME) evolve alongside genetic changes remains unclear.
This limits:
- The understanding of ccRCC evolution, as selection ultimately operates on phenotypes.
- Optimization of ICB timing, as the evolution of the TME remains poorly defined.
To fill this gap, we analysed 231 tumor samples from 79 ccRCC patients in the TRACERx Renal cohort.
By pairing DNA and RNA sequencing data, we could explore how the ccRCC TME and transcriptome evolve across multiple regions and disease stages.
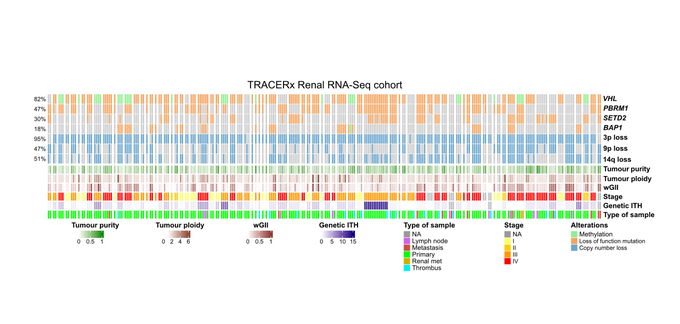
First, we found that cRCC shows pervasive non-genetic intratumor variation, which was only 50 percent explained by genetic heterogeneity.
This suggests a role for phenotypic plasticity in ccRCC evolution, as recently suggested in CRC.
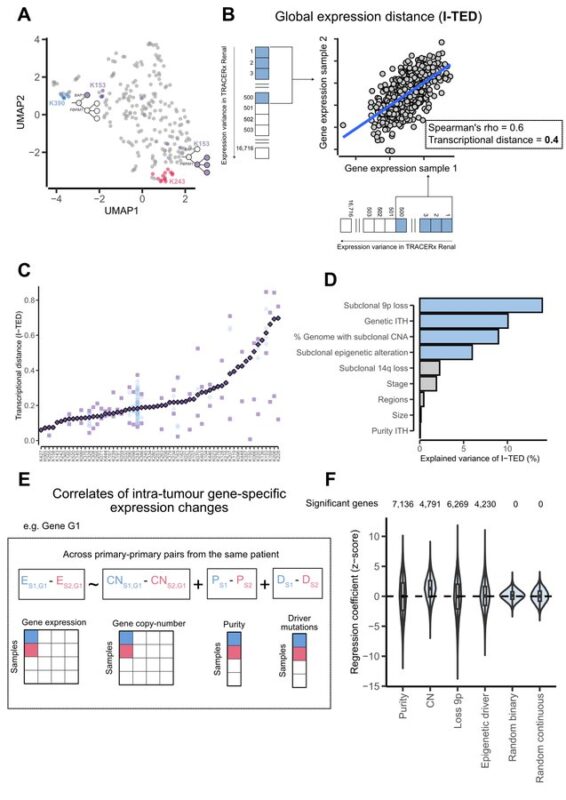
By integrating phylogenies with RNA-Seq data, we found recurrent patterns of ccRCC transcriptional evolution. Among others, we observed:
- Increased proliferation.
- Metabolic reprogramming.
These findings suggest convergent evolution to specific phenotypic states.
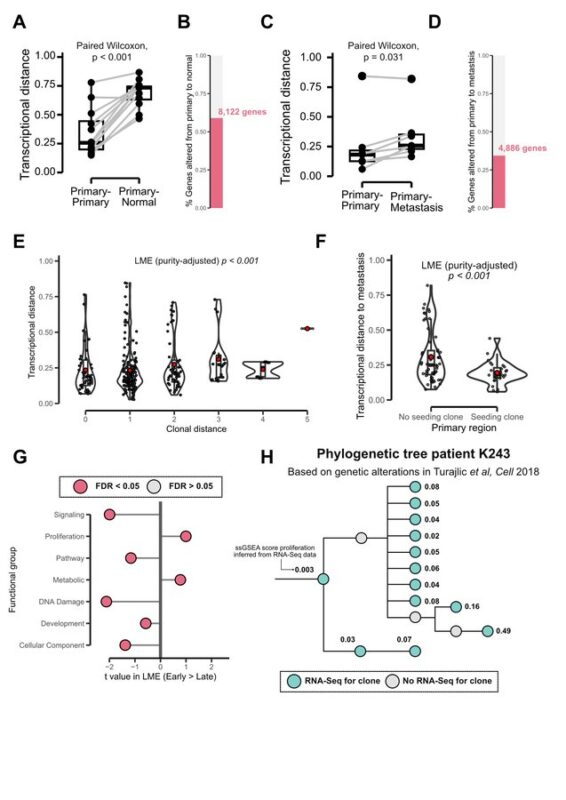
We explored the phenotypic impact of prognostic markers while controlling inter-patient variation:
- 9 patients’ loss co-occurred with higher proliferation and lower IFN-I expression.
- Aneuploidy occurred with overexpression of putative cGAS-STING repressors SLC19A1 and ENPP1.
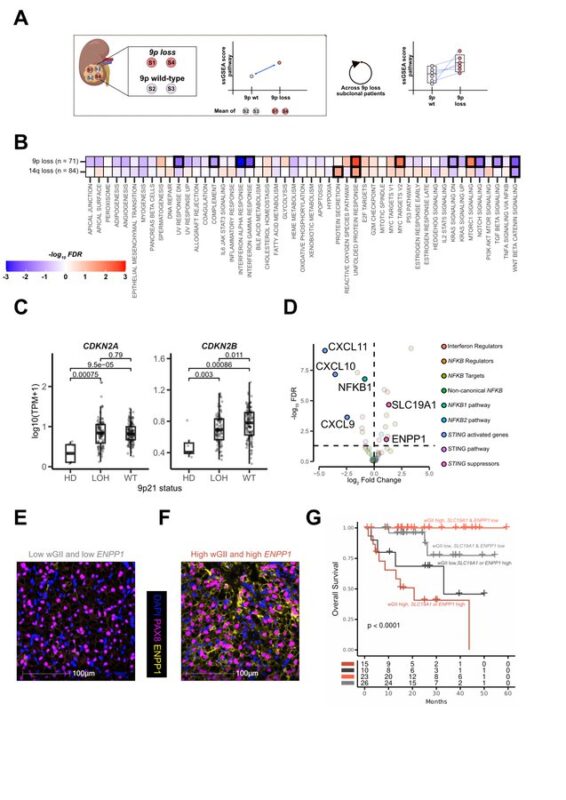
Overlaying transcriptional data enhanced the risk stratification provided by these two genetic biomarkers, suggesting that multi-modal biomarkers can better capture meaningful biology and refine ccRCC risk assessment.
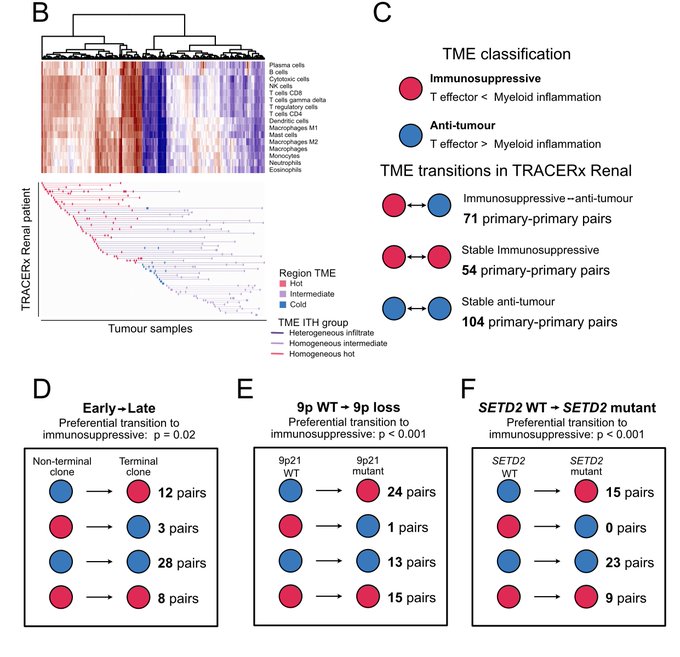
Through TME deconvolution of bulk RNA sequencing data, we identified an association between evolutionary trajectories and TME composition, suggesting a bidirectional relationship where genetic drivers of an evolutionary path influence TME composition and vice versa.
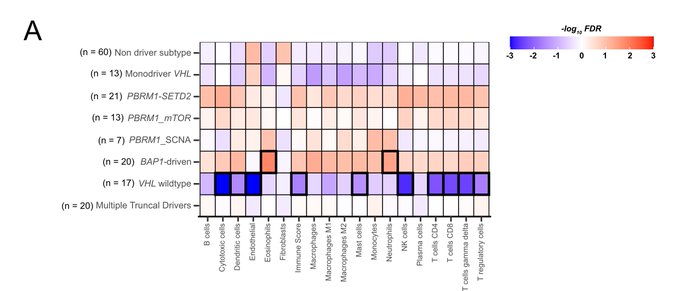
Combining tumour phylogenies and this TME inference, we identify progressive immune dysfunction during ccRCC evolution.
Shifts to anti-tumour TME recurrently co-occurred with 9 patients’ loss and SETD2 mutation.
This raises questions about the optimal timing of ICB.
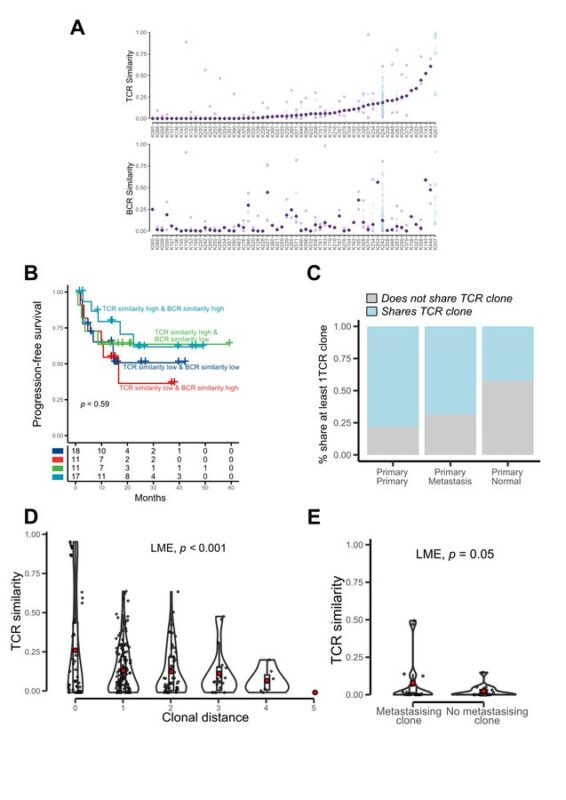
We also investigated TCR repertoire co-evolution with ccRCC:
TCR composition mirrored tumor clonal structure.
This persisted even in metastases, suggesting tumor-specific T cell clones may target heritable neoantigens acquired in primary tumors.
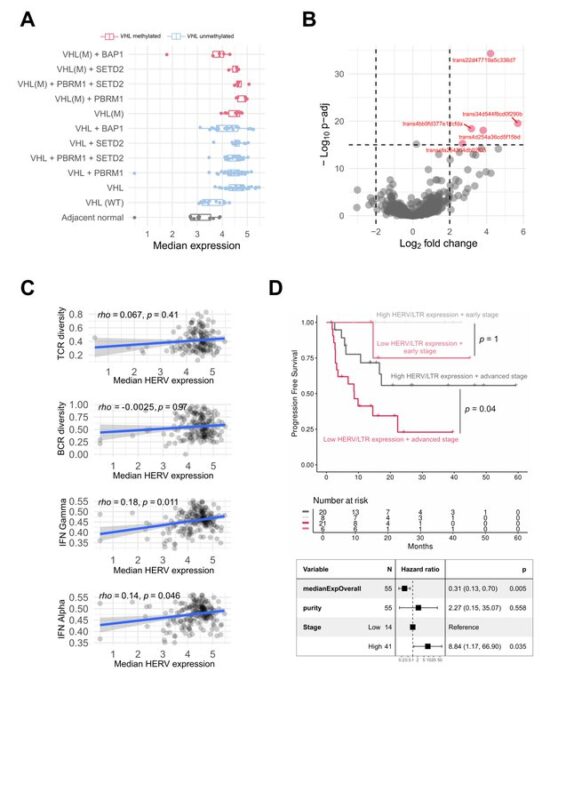
What about Human Endogenous Retroviruses (HERVs) as a source of neoantigens?
Early VHL LoF correlated with HERV de-repression, suggesting early occurrence.
However, HERV expression was not associated with cytotoxic immune infiltration nor TCR/BCR composition.
In conclusion, ccRCC evolution involves significant non-genetic variation that a solely genomic approach would miss.
Lots of more details to enjoy on the full story here!
Special kudos to Ángel Fernández Sanromán and Annika Fendler for leading this project to the end!
We’re deeply grateful to the patients and their families for participating, the clinical research teams, and our funders:
Science and Innovation at Cancer Research UK, BRCICR/RMH, and The Royal Marsden Cancer Charity.”
Tracking non-genetic evolution from primary to metastatic ccRCC: TRACERx Renal
Authors: Ángel Fernández-Sanromán, Annika Fendler, Benjy J.Y. Tan, Anne-Laure Cattin, Charlotte Spencer, Rachael Thompson, Lewis Au, Irene Lobon, Husayn Ahmed. Pallikonda, Alice Martin, Fiona Byrne, Antonia Franz, Anna Mikolajczak, Haseeb Rahman, Zayd Tippu, Scott T.C. Shepherd, Hugang Feng, Daqi Deng, Andrew Rowan, Lisa Pickering, Andrew J.S. Furness, Kate Young, David Nicol, Sarah Maria. Rudman, Tim O’Brien, Kim Edmonds, Ashish Chandra, Steve Hazell, Kevin Litchfield, George Kassiotis, James Larkin, Samra Turajlic.