Yu-Hung Wang, PhD student at The University of Manchester, shared on X
“Delighted to share our latest story!
Prognostic and therapeutic implications of TP53 expression in chronic myelomonocytic leukemia(CMML).

We first corroborated the paucity of TP53 mutations in CMML across previously unreported UK and Taiwan cohorts. As expected, UK TP53-altered patients displayed significantly inferior outcomes compared with TP53 WT patients, with a similar trend in the Taiwan cohort.

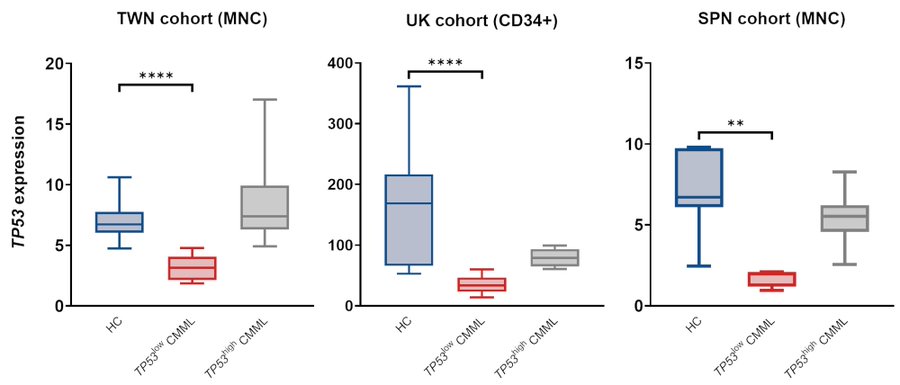
In our Taiwan RNA-sequencing cohort, TP53-low patients displayed significantly lower expression than healthy controls, while TP53-high expression levels were comparable to HCs. Thus, while most CMML pts are TP53 WT, BM cells from a subset display abnormally low TP53 expression.
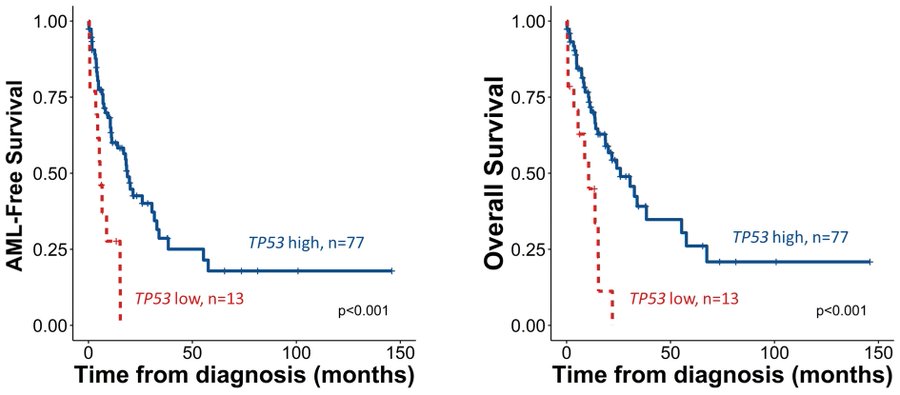
TP53-low patients displayed significantly shorter LFS and OS than TP53-high cases. In subgroup analyses, lower TP53 retained strong predictive value for LFS and OS even within CPSS and CPSS-Mol-stratified subgroups, and in exclusively ASXL1 WT patients.
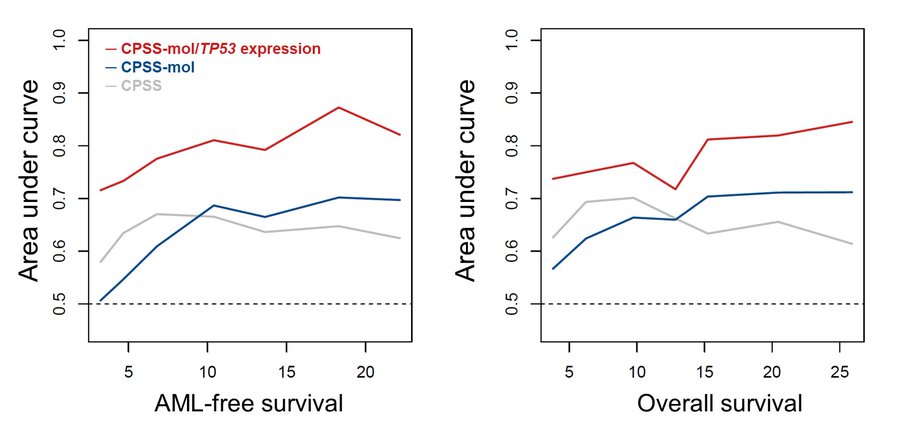
Time-dependent ROC curve analysis revealed potential for TP53 expression to enhance current prognostication systems. In multivariable analysis, lower TP53 expression remained prognostically detrimental for LFS and OS.
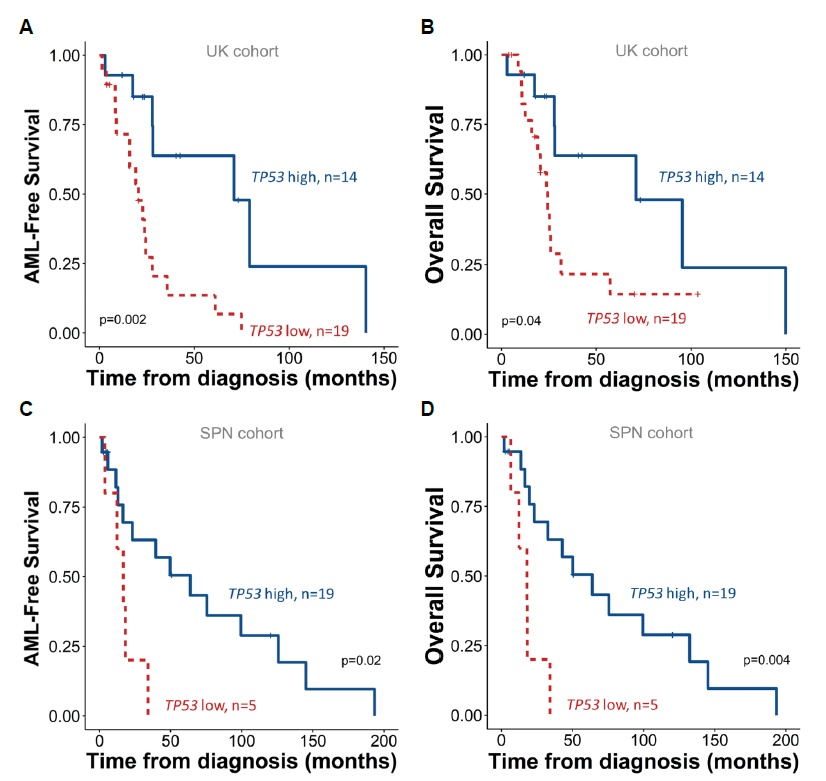
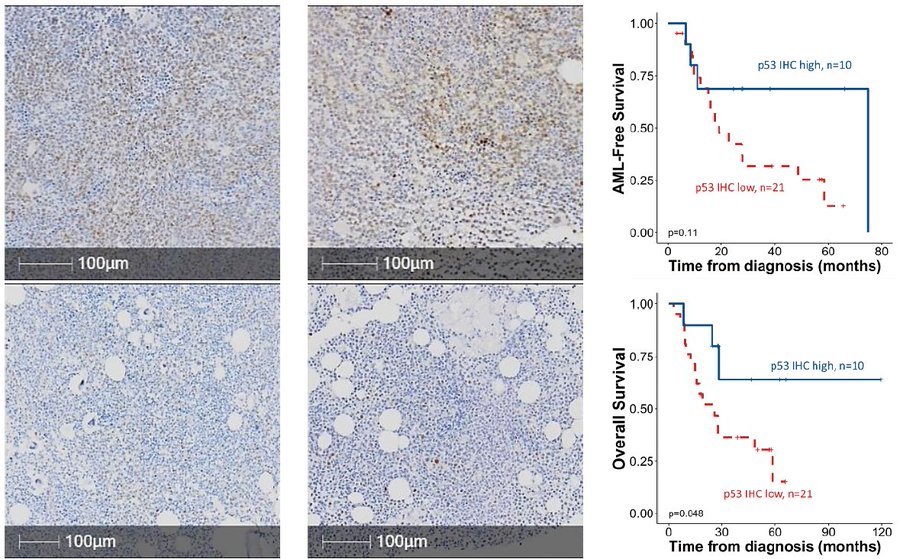
This was consistent across validation cohorts, strengthening the observed link between lower TP53 expression and adverse outcomes.


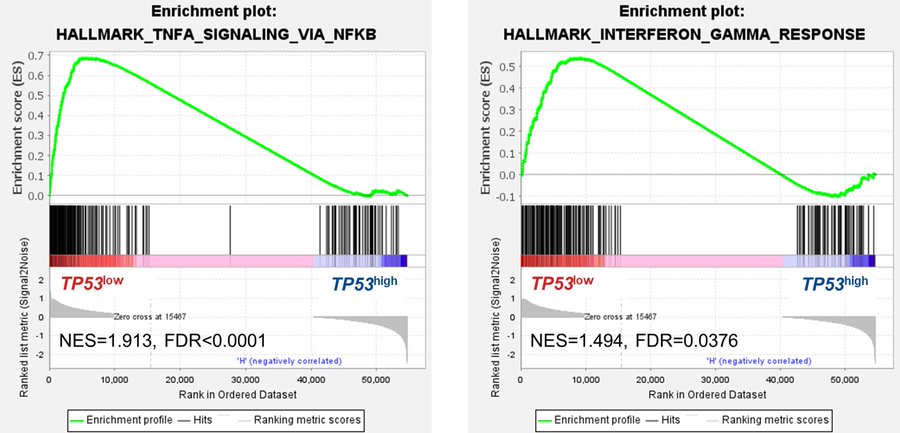
Interestingly, these were all among the most upregulated pathways in TP53-MT (vs TP53 WT) samples across multiple cancers in TCGA data, implying that the driving biology of TP53low CMML is distinct from (and not functionally equivalent to) that of oncogenic TP53 mutations.
Conversely, TP53-low patients demonstrated enhanced TNF-alpha and inflammatory signals, highlighting possible crosstalk between p53 and extrinsic factors in the CMML BM microenvironment.
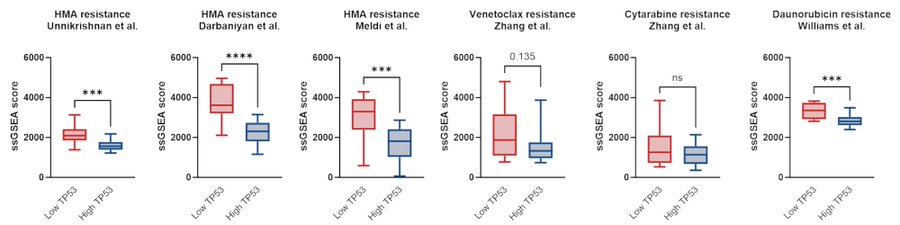
Similar results were observed in our other cohorts. Taken together, compared with HCs or TP53-high CMML, TP53-low CMML cells display relatively quiescent cell cycle but heightened inflammation.
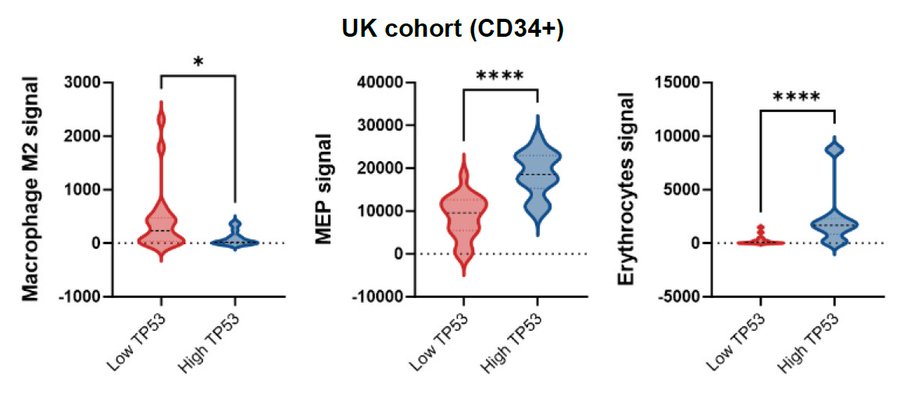
With emerging evidence suggesting discrete roles for p53 in regulating inflammation and immune cell landscape, we applied xCell to analyze signals of 22 cell types. TP53-low CMML displayed stronger M2-macrophage, but lower megakaryocytic-erythroid progenitors (MEP) signals.

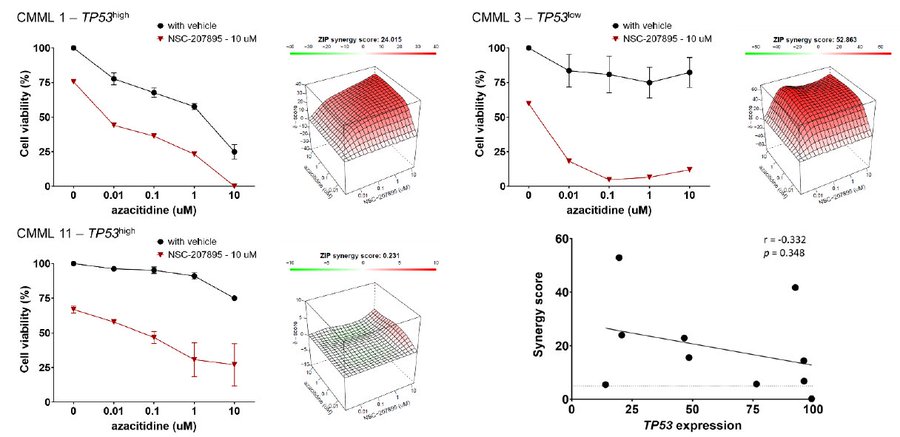
Finally, we explored whether reduced TP53 expression in this CMML subset could be exploited therapeutically. Combining NSC-207895, a dual MDMX/MDM2 inhibitor, and p53 activator, with AZA at various concentrations, we observed clear and substantial synergy in 10/11 primary samples.
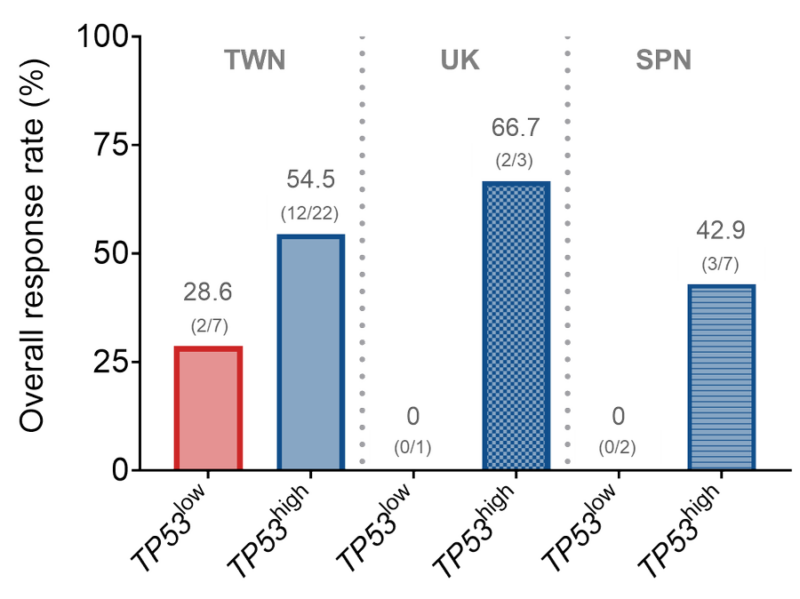
There was a trend towards inverse correlation between TP53 expression and empirical synergy scores, suggesting potential for pharmacological p53 activation to enhance HMA sensitivity in CMML with broad efficacy; perhaps preferentially in adverse TP53-low expressing cases.
An intriguing question remains: why are TP53 mutations so infrequent in CMML? Speculatively, they might cause unknown synthetic lethalities in CMML cells or promote alternative lineage pathways, altering disease classification.
We observed significant under-representation of myelomonocytic/blastic M4/M5 FAB subtypes associated with TP53 mutations amongst 1511 AML cases at NTUH, and re-analyzing 577 cases from TCGA and BeatAML datasets (odds ratio 0.48 and 0.49, respectively).
Thus, acquisition of TP53 mutations onto the canonical CMML mutation background might alter the resultant phenotype away from clinicopathological features compatible with CMML diagnostic criteria.
In conclusion, our study is the first to link low TP53 expression with distinct features and outcomes in CMML. We confirm the rarity of TP53 mutations and identify a novel subgroup with low TP53 expression, higher HMA resistance, unique biology, and poorer prognosis.
We highlight potential for combining HMA and MDMX/MDM2 inhibition to restore HMA sensitivity, as an attractive candidate therapeutic approach for clinical study to address this unmet clinical need.
Finally, my sincere appreciation to patients, my supervisors, colleagues, friends, and collaborators.”
Read Further.
Source: Yu-Hung Wang/X