In a landmark study published in Nature Cell Biology, Neville et al. unveil a previously unrecognized epigenetic mechanism that reshapes our understanding of transcriptional regulation in leukemia. The research demonstrates that DOT1L, a histone methyltransferase long associated with MLL-rearranged leukemia, serves as a molecular guardian of transcriptional memory by directly antagonizing PRC1.1, a non-canonical Polycomb repressive complex.
This discovery does more than clarify the biology of MLL-fusion leukemias it reframes DOT1L as a central regulator balancing activation and repression within the broader MLL–Polycomb axis.
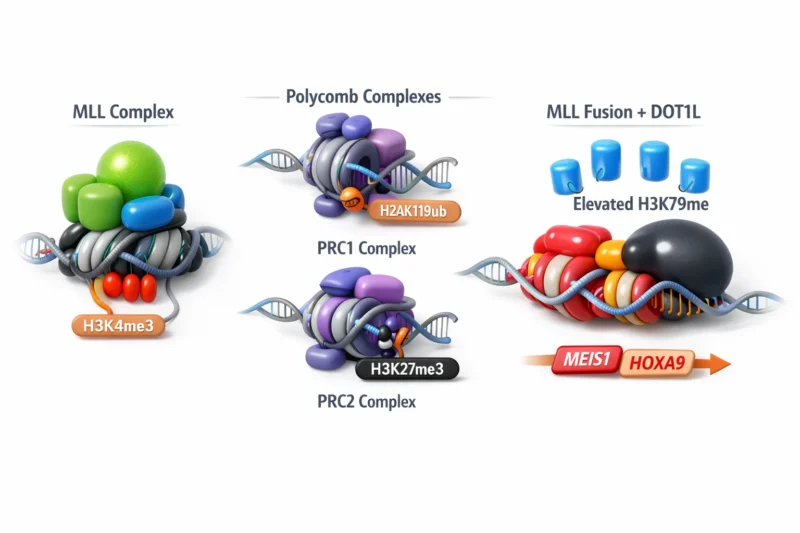
The MLL–Polycomb Axis: A Delicate Epigenetic Balance
Normal gene regulation depends on a dynamic equilibrium between activating and repressive chromatin modifiers.
- MLL complexes deposit H3K4me3, promoting transcriptional activation.
- Polycomb complexes (PRC1 and PRC2) deposit repressive marks such as H2AK119ub and H3K27me3, enforcing gene silencing.
In MLL-rearranged leukemia, oncogenic MLL fusion proteins aberrantly recruit DOT1L, leading to elevated H3K79 methylation at key leukemogenic genes like MEIS1 and HOXA9. While DOT1L inhibitors have been clinically explored, their precise mechanistic impact remained unclear until now.
PRC1.1: The Missing Mediator of Therapeutic Response
Using genome-wide CRISPR screens, transcriptomic profiling, and chromatin immunoprecipitation sequencing (ChIP-seq), the authors identified PRC1.1 components—particularly BCOR and PCGF1—as essential mediators of both DOT1L and Menin inhibitor efficacy.
Key findings include:
- Loss of PRC1.1 components confers resistance to DOT1L and Menin inhibition.
- Therapeutic gene repression requires PRC1.1-mediated deposition of H2AK119ub.
- Without PRC1.1, leukemic cells fail to undergo proper differentiation, both in vitro and in vivo.
Importantly, these results demonstrate that DOT1L and Menin inhibitors do not act solely by suppressing transcriptional elongation. Instead, their therapeutic success depends on enabling Polycomb-mediated repression.
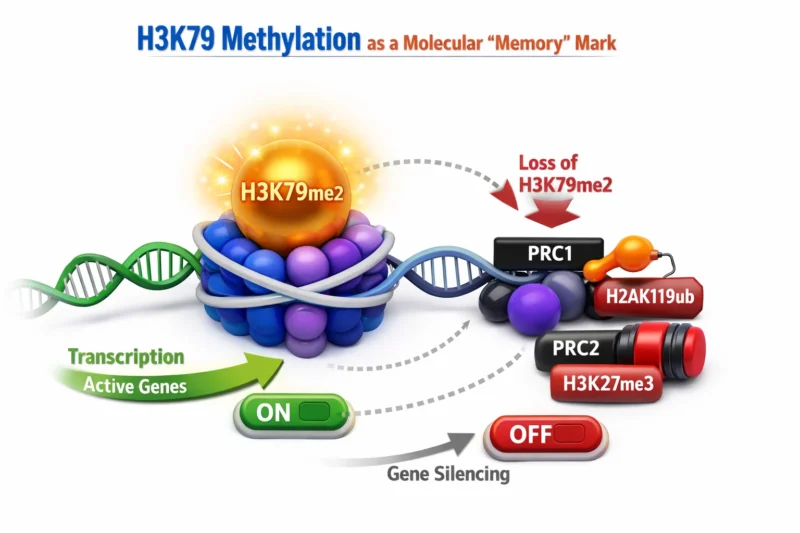
H3K79 Methylation as a Molecular “Memory” Mark
The study’s most transformative insight is the identification of H3K79 methylation as a transcriptional memory mark.
Unlike many histone modifications, H3K79me2 lacks a dedicated demethylase. Its slow turnover means that once deposited, it persists across cell divisions. The authors show that this stability allows H3K79me2 to protect active genes from premature Polycomb repression.
Mechanistically:
- Loss of H3K79me2 strongly correlates with increased H2AK119ub deposition.
- In vitro assays confirm that H3K79me2/3 directly inhibits the catalytic activity of PRC1 complexes.
- Menin inhibition initially causes reversible gene repression. Only after H3K79me2 decays does irreversible Polycomb silencing occur.
This reveals a stepwise epigenetic switch:
- Menin inhibition displaces MLL-fusion complexes.
- H3K79me2 gradually declines.
- PRC1.1 deposits H2AK119ub.
- PRC2 establishes stable H3K27me3-mediated repression.
The delay introduced by H3K79 methylation acts as a biological buffer, preventing transient transcriptional fluctuations from triggering permanent silencing.
Irreversible Repression and Therapeutic Implications
One of the study’s most clinically relevant findings is that prolonged Menin inhibition induces irreversible Polycomb-mediated repression, even after drug withdrawal.
This suggests:
- Short, intensive dosing schedules may achieve durable responses.
- Transient Menin inhibition can permanently reprogram leukemic cells.
- PRC1.1 integrity is essential for long-term therapeutic benefit.
These insights may help explain why Menin inhibitors—recently approved for relapsed MLL-rearranged and NPM1-mutant AML—have demonstrated stronger clinical efficacy than DOT1L inhibitors.
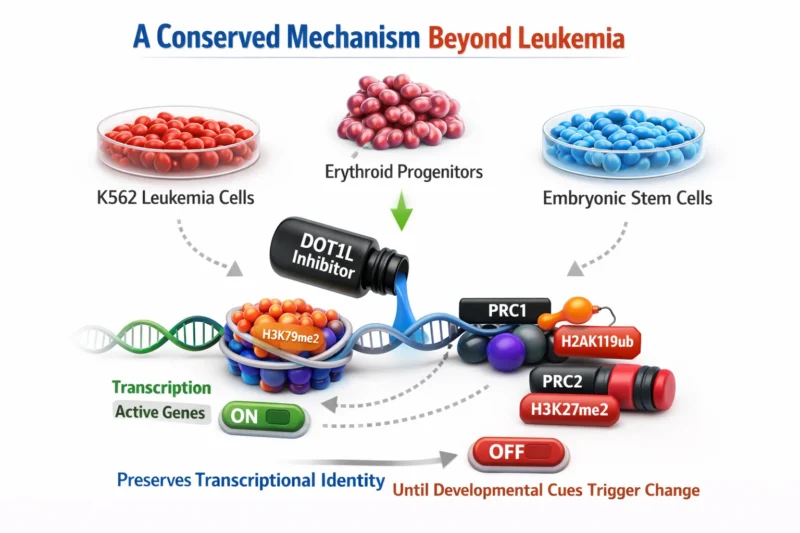
A Conserved Mechanism Beyond Leukemia
Strikingly, DOT1L–PRC1 antagonism is not restricted to MLL-rearranged leukemia.
Across multiple cell types—including K562 cells, erythroid progenitors, and mouse embryonic stem cells—DOT1L inhibition globally reduced H3K79me2 and modestly increased H2AK119ub.
Rather than causing widespread gene silencing, this shift appears to “prime” cells for differentiation. In erythroid progenitors, DOT1L inhibition accelerated differentiation-associated repression, reinforcing the concept that DOT1L preserves transcriptional identity until developmental cues trigger change.
A New Model of Epigenetic Memory
This work proposes a unifying model:
- H3K79me2 encodes memory of gene activation.
- H3K27me3 encodes memory of repression.
- The balance between these marks stabilizes cell identity.
By hijacking DOT1L, MLL-fusion proteins exploit this conserved buffering system to maintain leukemogenic gene expression.
Therapeutically, disrupting this balance allows Polycomb complexes to reassert control—converting transient inhibition into permanent epigenetic reprogramming.
Written by Nare Hovhannisyan, MD


