The c-TRAK TN Trial , presented at the 2025 San Antonio Breast Cancer Symposium (SABCS), reported results from a tissue-free circulating tumor DNA (ctDNA) analysis, providing new evidence that methylation-based, tumor-agnostic assays detect minimal residual disease earlier and in more patients with high-risk early triple-negative breast cancer than tumor-informed approaches.
Background and Rationale
Detection of ctDNA after completion of curative-intent therapy is a powerful prognostic marker for recurrence in early breast cancer. To date, most supporting evidence has come from tumor-informed assays, which require sequencing of the primary tumor to design personalized mutation panels. While analytically sensitive, these approaches are limited by the need for archival tissue, sequencing turnaround time, and logistical complexity.
Tissue-free ctDNA assays, which do not rely on prior tumor sequencing, offer a potential alternative if sufficient accuracy and prognostic performance can be demonstrated. The current analysis aimed to evaluate the prognostic significance of tissue-free ctDNA detection in early TNBC and to directly compare its performance with tumor-informed digital droplet PCR (ddPCR).
c-TRAK TN Trial’s Study Design
The analysis utilized plasma samples from c-TRAK TN, the first prospective ctDNA surveillance study in early-stage TNBC. In c-TRAK TN, patients at moderate to high risk of relapse underwent serial ctDNA testing every three months following completion of standard therapy, using ddPCR to track tumor-specific mutations.
Archived plasma samples from this cohort were retrospectively analyzed using a tissue-free, methylation-based ctDNA assay, enabling a direct comparison between approaches within the same well-characterized population.
Tissue-Free ctDNA Assay Methodology
The tissue-free assay exploits differentially methylated regions (DMRs) between cancer and non-cancer DNA. Cell-free DNA was extracted from 2–4 mL of plasma, partitioned by methylation status, and enriched using a targeted capture panel covering approximately 20,000 DMRs, including 3,000 breast-specific regions. Tumor methylation fraction was reported in ctDNA-positive samples and correlated with tumor purity.
Importantly, this approach eliminates the need for tumor sequencing while maintaining biological specificity for breast cancer–derived ctDNA.
Patient Cohort and Follow-Up
A total of 1,026 plasma samples from 159 patients were analyzed using the tissue-free assay, with a median of 10 samples per patient. Quality control pass rates were high (98.6%). Median follow-up from the first surveillance blood draw was 33.9 months.
The cohort was representative of high-risk early TNBC, with the majority of tumors being high grade and most patients having received neoadjuvant and/or adjuvant chemotherapy.
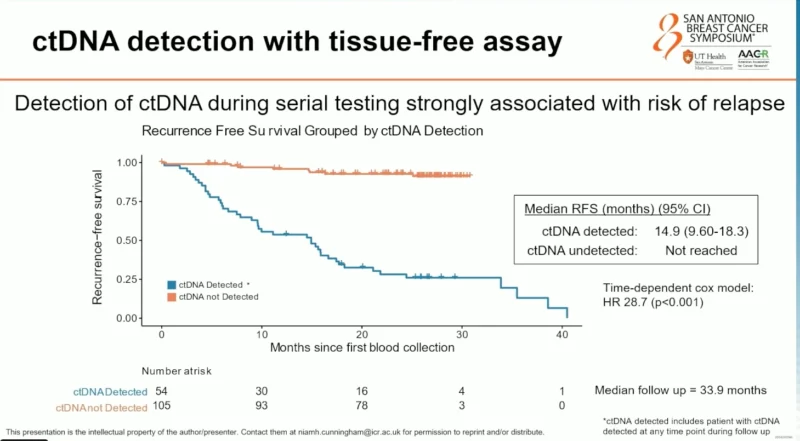
ctDNA Detection and Risk of Recurrence
Detection of ctDNA at any time point during serial surveillance was strongly associated with recurrence risk. Patients with detectable ctDNA had a median recurrence-free survival (RFS) of 14.9 months, whereas median RFS was not reached in patients without ctDNA detection.
The association was robust, with a time-dependent hazard ratio of 28.7 (p < 0.001), underscoring the powerful prognostic value of ctDNA detection in this high-risk population.
Tissue-Free Assay Versus Tumor-Informed ddPCR
When directly compared with ddPCR, the tissue-free assay demonstrated 95.4% concordance at the sample level. Among 42 patients with ctDNA detected by both methods at any time point, two-thirds were detected simultaneously by both assays. Notably, 33.3% were detected earlier by the tissue-free assay, while no patients were detected earlier by ddPCR.
At 12 months, the estimated ctDNA detection rate was 29.0% with the tissue-free assay compared with 23.7% with ddPCR, indicating that the tissue-free approach identified ctDNA in more patients.
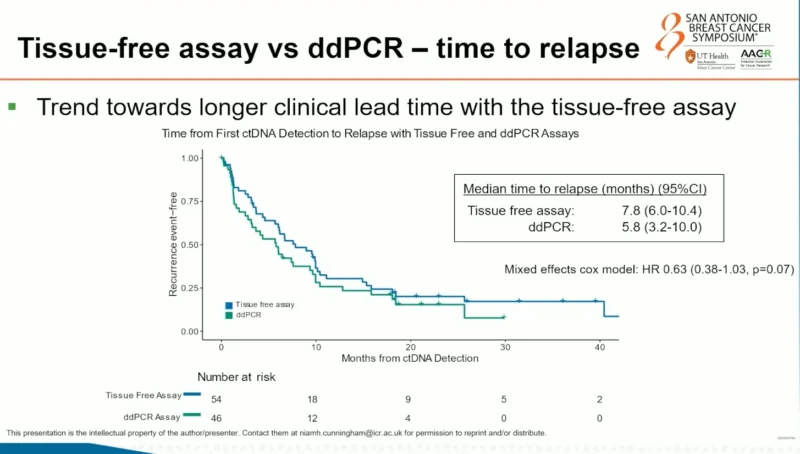
Clinical Lead Time to Relapse
The tissue-free assay also showed a trend toward a longer clinical lead time between ctDNA detection and overt relapse. Median time from first ctDNA detection to recurrence was 7.8 months with the tissue-free assay versus 5.8 months with ddPCR. Although this difference did not reach conventional statistical significance (HR 0.63, p = 0.07), the numerical advantage suggests earlier molecular detection may be achievable without tumor sequencing.
Conclusions and Implications
This SABCS 2025 presentation demonstrates that tissue-free ctDNA detection can anticipate relapse with high accuracy in patients with high-risk early TNBC. Compared with tumor-informed ddPCR, the tissue-free assay detected ctDNA more frequently and at earlier time points, while maintaining strong prognostic discrimination.
Ongoing comparisons with whole-exome sequencing–based tumor-informed assays will further clarify relative performance. While these findings establish strong analytical and prognostic validity, prospective interventional studies will be required to determine whether tissue-free ctDNA surveillance can guide treatment decisions and improve clinical outcomes.
For more information click here.


