ctDNA (Circulating tumor DNA) has emerged as a key tool for detecting molecular residual disease after curative surgery in early-stage colorectal cancer, often identifying recurrence months before radiologic relapse and informing adjuvant and surveillance strategies. However, clinicians must choose between tumor-informed assays, which are personalized and potentially more sensitive, and tumor-agnostic assays, which are faster and tissue-independent but may miss low-level disease.
In this diagnostic accuracy meta-analysis, Camblor and colleagues show that this choice is highly context-dependent: during serial monitoring, tumor-informed assays achieve significantly higher sensitivity than tumor-agnostic approaches without increasing false positives, underscoring the importance of aligning ctDNA testing strategy with clinical intent.
Title: Clinical performance of tumor-informed versus tumor-agnostic ctDNA assays for colorectal cancer recurrence: A systematic review and diagnostic accuracy meta-analysis
Authors: Daniel G. Camblor Belén Martínez-Castedo Jorge Martín-Arana Francisco Gimeno-Valiente Blanca García-Micó Francisco Martínez-Picó Víctor Segu Miguel García-Bartolomé Diego González Alejandro Guimera Marisol Huerta Susana Roselló Valentina Gambardella Desamparados Roda Leontios Pappas Aparna Parikh Juan Antonio Carbonell-Asins Andrés Cervantes Noelia Tarazona Show less
Background
After curative-intent surgery for stage I–III colorectal cancer (CRC), the central clinical problem is identifying who still has molecular residual disease (MRD) and is therefore at high risk of recurrence—often months before imaging becomes positive. Circulating tumor DNA (ctDNA) has emerged as a practical MRD tool to refine adjuvant chemotherapy decisions and improve surveillance, but clinicians face a real-world choice between tumor-informed (TI) assays (personalized to the patient’s tumor mutations) and tumor-agnostic (TA) assays (plasma-only or fixed panels, often including methylation/fragmentomic strategies) that are easier to deploy when tissue is limited.
This systematic review and diagnostic accuracy meta-analysis directly compared TI vs TA performance for detecting recurrence and, critically, tested how sampling strategy (a single postoperative “landmark” test vs repeated “serial” testing) changes sensitivity and false-positive rates in practice.
Methods
The authors performed a systematic search of PubMed and Embase through October 31, 2025, restricted to English-language full texts from 2015 onward, focusing on resected CRC in the postoperative setting and requiring extractable 2×2 diagnostic accuracy data (true/false positives/negatives) for recurrence detection. Study reporting followed PRISMA-DTA, and risk of bias was assessed using QUADAS-2. For quantitative synthesis, they used (1) a univariate random-effects approach to explore diagnostic odds ratios (DORs) and publication bias (Deeks’ test), and (2) the Reitsma bivariate random-effects model to jointly estimate sensitivity and false-positive rate (FPR = 1–specificity) and to compare TI vs TA within landmark and serial settings.
Study Design
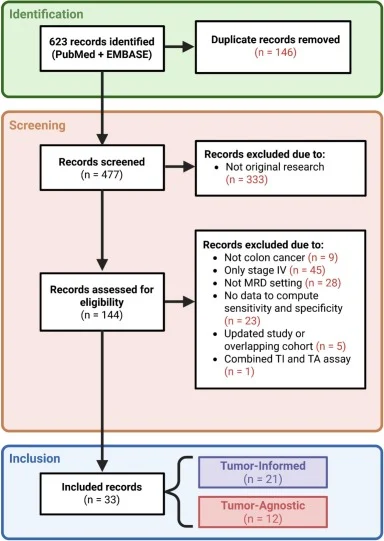
This was a systematic review with diagnostic accuracy meta-analysis (not a therapeutic trial). Across all eligible publications, the authors included 33 studies evaluating ctDNA for recurrence detection after curative-intent CRC surgery. Real-world implementation was explicitly addressed by stratifying studies into: Landmark testing (single postoperative timepoint, typically weeks after surgery) versus Serial testing (longitudinal sampling across follow-up). The primary comparison was TI vs TA, and the major design question was whether assay type matters more—or less—than sampling strategy.
Results
Study volume and clinical scenarios represented. From 623 records, 33 studies were included. For landmark analyses, 27 studies contributed data from 7,482 patients with 1,488 recurrence events. For serial analyses, 17 studies included 2,865 patients with 561 recurrences, reflecting fewer datasets but closer alignment with how ctDNA is often used for ongoing surveillance.
Sampling strategy changed sensitivity more than it changed false positives. In the bivariate framework, serial testing improved sensitivity compared with landmark-only testing (0.72 vs 0.51, p = 0.001), while differences in false-positive rates were not statistically significant between strategies. This supports a clinically intuitive point: repeated sampling increases the chance of catching ctDNA as tumor shedding evolves over time, without clearly increasing the burden of false alarms.
Tumor-informed vs tumor-agnostic—where the difference becomes decisive. When the analysis was stratified by setting, the major separation appeared in the serial scenario. In serial monitoring, TI assays showed markedly higher sensitivity than TA assays (0.88 vs 0.59, p = 0.001). Importantly, this gain did not come with a statistically higher false-positive rate: FPR was 0.09 for TI vs 0.12 for TA (p = 0.419). In other words, serial TI testing was substantially better at identifying true recurrences while maintaining a similar false-positive profile.
In landmark (single timepoint) testing, TI did not outperform TA. In landmark-only studies, sensitivity estimates were 0.48 for TI vs 0.58 for TA (p = 0.070), and FPRs were similar (0.08 vs 0.10, p = 0.329). Practically, this suggests that when only a one-time postoperative ctDNA result is feasible—often the scenario tied to time-sensitive adjuvant decisions—assay selection may hinge more on logistics (tissue availability, turnaround time) than on an expectation of clearly superior diagnostic accuracy from one approach.
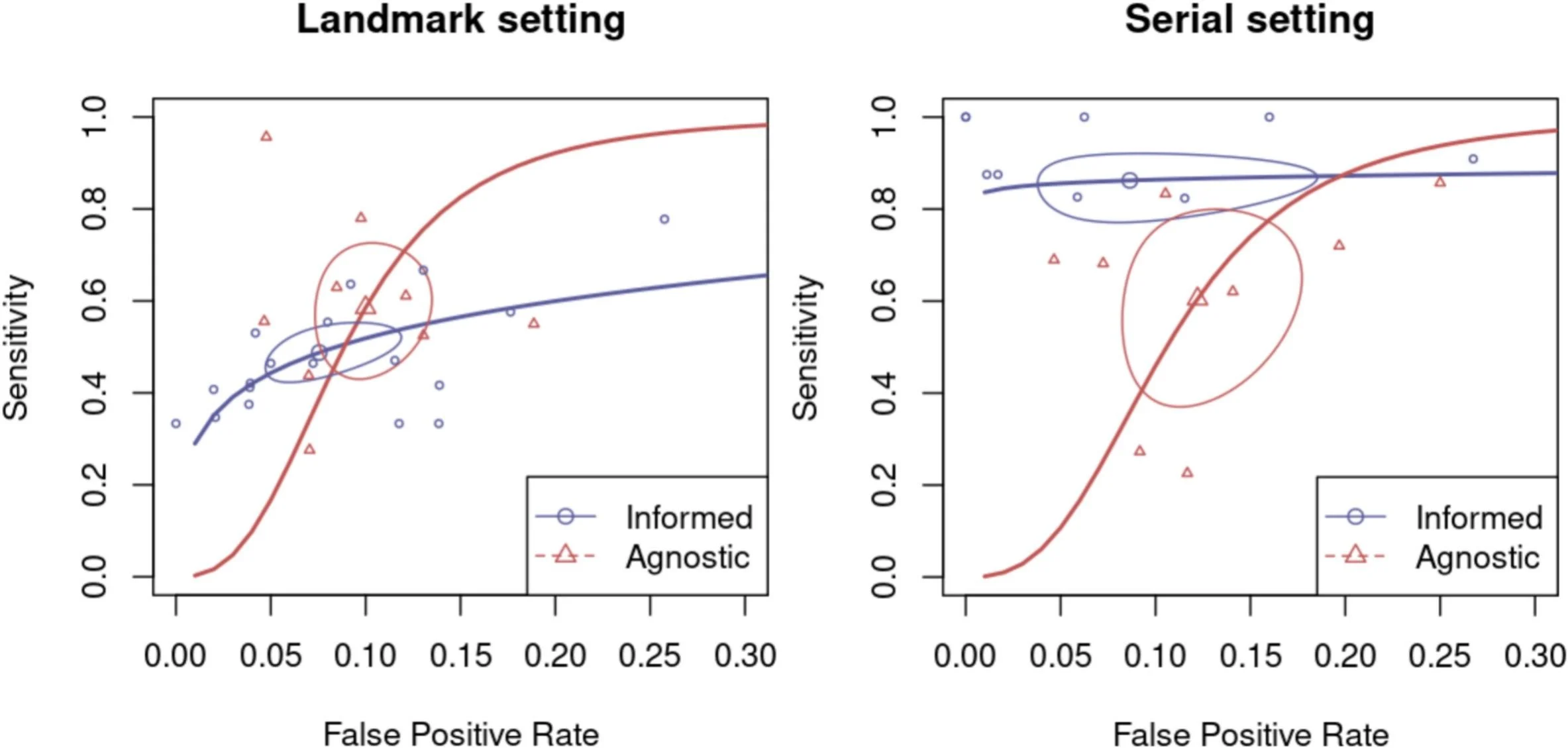
Exploratory DOR signals and publication bias. In univariate analyses, landmark studies showed similar pooled DORs for TI and TA, whereas serial studies favored TI with a much higher pooled DOR signal. However, Deeks’ test suggested publication bias in the serial dataset (smaller studies tending to “overperform”), which the authors appropriately flag as a caution when interpreting the magnitude of advantage—without negating the consistent directionality of better sensitivity for TI in serial monitoring.
Key Findings
- Serial sampling meaningfully improves recurrence detection sensitivity compared with a single landmark test, reinforcing that how often you test materially affects ctDNA performance.
- In serial surveillance, tumor-informed ctDNA is substantially more sensitive than tumor-agnostic approaches (0.88 vs 0.59) while maintaining similar false-positive rates, supporting TI as the more reliable option when longitudinal monitoring is feasible.
- In the landmark postoperative window, no statistically significant diagnostic advantage was demonstrated for TI over TA, which keeps TA approaches clinically relevant when tumor tissue is unavailable or decisions must be made quickly.
Conclusion
This diagnostic accuracy meta-analysis provides a clinically practical framework for selecting ctDNA assays after CRC resection: sampling strategy strongly influences performance, and tumor-informed testing clearly strengthens sensitivity in serial monitoring (0.88 vs 0.59) without significantly worsening false-positive rates. In contrast, when ctDNA is used as a single landmark postoperative test, TI does not demonstrate a statistically significant advantage over TA, making real-world constraints—particularly tumor tissue availability and timing—central to decision-making.
You can read the full article here.