Mainak Bardhan, Doctoral Researcher at Howard Hughes Medical Institute-University of Miami Miller School of Medicine, shared a post by Avash Das, Resident Doctor at Harvard Medical School, adding:
“Exciting thread on blood cancer by Avash Das from Harvard Medical School.”
Quoting Avash Das‘s post:
“Taking T-Acute Lymphoblastic Leukemia (T-ALL) a ‘NOTCH’ higher (1/7): T-ALL is a hematological malignancy characterized by the infiltration of bone marrow with hematopoietic cells expressing immature T-cell markers. It constitutes approximately 15% of pediatric and 25% of adult newly diagnosed ALL cases and can have a poor prognosis from relapse or refractory disease. Approximately 50-60% of T-ALL cases can be causally linked to a gain-of-function mutation in NOTCH1.
NOTCH1 is a class I transmembrane glycoprotein essential for T cell maturation. It drives the commitment of common lymphoid progenitors to the T cell lineage by promoting T cell-specific gene expression and suppressing alternative cell fates and differentiation.
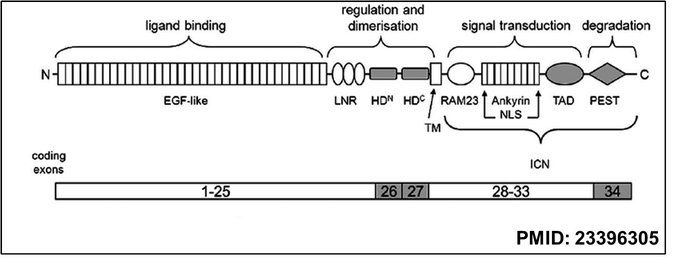
Upon examining the domain architecture of the NOTCH1 receptor protein, with functional regions annotated across its N-terminal to C-terminal ends, it shows EGF-like repeats for ligand binding, LNR and HD domains for regulation and dimerisation, and RAM23, Ankyrin, TAD, and PEST domains for signal transduction and degradation. The mapped exonic organisation indicates that exons 1–25 encode the extracellular domain, exons 26–27 span the transmembrane region, and exons 28–34 encode the intracellular domain (ICN) that mediates transcriptional activation after cleavage.
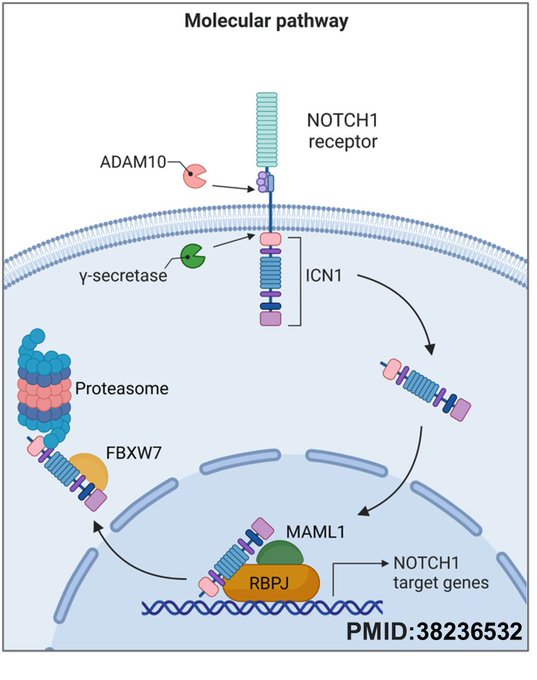
The canonical NOTCH1 signalling pathway functions in the following sequence: the ligand binding induces proteolytic cleavage of the NOTCH1 receptor by ADAM10 and γ-secretase, releasing the intracellular domain (ICN1). ICN1 translocate to the nucleus, forms a transcriptional activation complex with RBPJ and MAML1 to drive expression of NOTCH1 target genes, and is ultimately ubiquitinated by FBXW7 and degraded by the proteasome to terminate signalling.
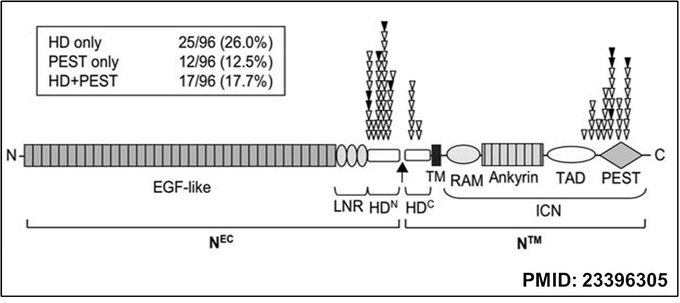
The most common mutation sites in NOTCH1 in T-cell acute lymphoblastic leukemia (T-ALL) are in the heterodimerization (HD) domain (26%) and the C-terminal PEST domain (12.5%) or both (17.7%). The most common mutations are missense mutations, small in-frame insertions or deletions for the HD domain, and frameshift or nonsense mutations for the PEST domain. NOTCH1 HD inactivating mutations cause ligand-independent activation of the receptor. In contrast, PEST domain mutations cause ligand-independent activation and impair proteasomal degradation of the intracellular NOTCH1, leading to sustained transcription of NOTCH1 target genes and contributing to leukemogenesis.
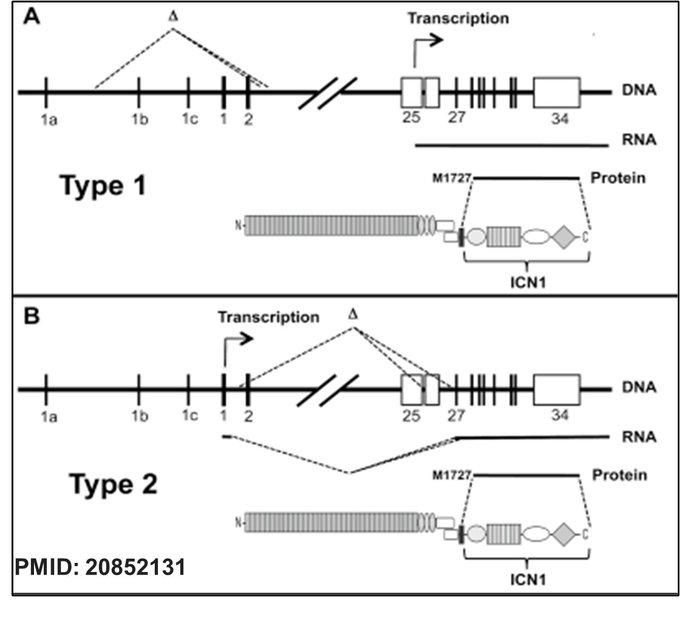
Additionally, Aster and colleagues have also described the rare incidence of intragenic deletion of NOTCH1 in T-ALL (PMID: 20852131). Two types of intergenic deletions in NOTCH1 leading to aberrant ICN1 expression T-ALL have been reported: In Type 1, a deletion upstream of exon 25 removes regulatory elements and juxtaposes cryptic promoter elements to exon 25, initiating transcription at exon 25; in Type 2, transcription starts aberrantly at exon 1 or 2 but splices directly into exon 25 due to a deletion, both ultimately producing a truncated NOTCH1 protein that includes the active ICN1 domain.
Identifying NOTCH1 mutations or intragenic deletions is important for diagnostic and prognostic purposes. As the key driver of leukemogenesis in T-ALL, NOTCH1 mutations can clinch the diagnosis. While NOTCH1 mutations may not define prognosis, they can serve as a molecular marker for minimal residual disease (MRD) in selected patients. Emerging therapies like γ-Secretase inhibitors (GSIs) and combination therapies with glucocorticoids, mTOR inhibitors, or BCL2 inhibitors, BET inhibitors and CDK7 inhibitors are currently being tested; there are no FDA-approved targeted therapies for NOTCH1 in T-ALL.
On a personal note, we had a patient with suspected T-ALL who had a negative NOTCH1 variant in our hybrid capture-based targeted Next Generation Sequencing (NGS) assay, but we were able to detect an intragenic NOTCH1 deletion between intron 2 and exon 28 in an amplicon-based NGS assay dedicated to identifying fusion partners, highlighting the utility of various NGS panel based assays in molecular and precision diagnostics as well.”
More posts featuring Mainak Bardhan.


