Radhika Mathur, Research Scientist at UCSF, recently shared on X/Twitter:
“Thrilled to finally share my 5+ years of postdoctoral work, published today in Cell!
‘Glioblastoma evolution and heterogeneity from a 3D whole-tumor perspective.’
The brain tumor glioblastoma remains incurable. Treatment failure is often attributed to intratumoral heterogeneity, but this is difficult to study as typically only one sample is acquired per patient.
We worked with a clinical team at the Department of Neurological Surgery including Shawn Hervey-Jumper to design a new 3D spatial sampling strategy. We used 3D surgical neuronavigation to acquire 10+ samples per patient, each mapped by 3D spatial coordinates and integrated these with patient MRI scans.

We generated 3D models revealing sample locations for each patient in the context of the whole tumor. We then subjected spatially mapped samples to integrative genomic and epigenomic assays including whole-exome, RNA-Seq, ATAC-Seq (including at the single-nucleus level), and Hi-C.

We distinguished molecular features and programs present tumor-wide from those with regional specificity, allowing us to infer GBM evolutionary trajectories. We revealed new insights on GBM cell-of-origin and identified chromothripsis as a frequent early driver event.
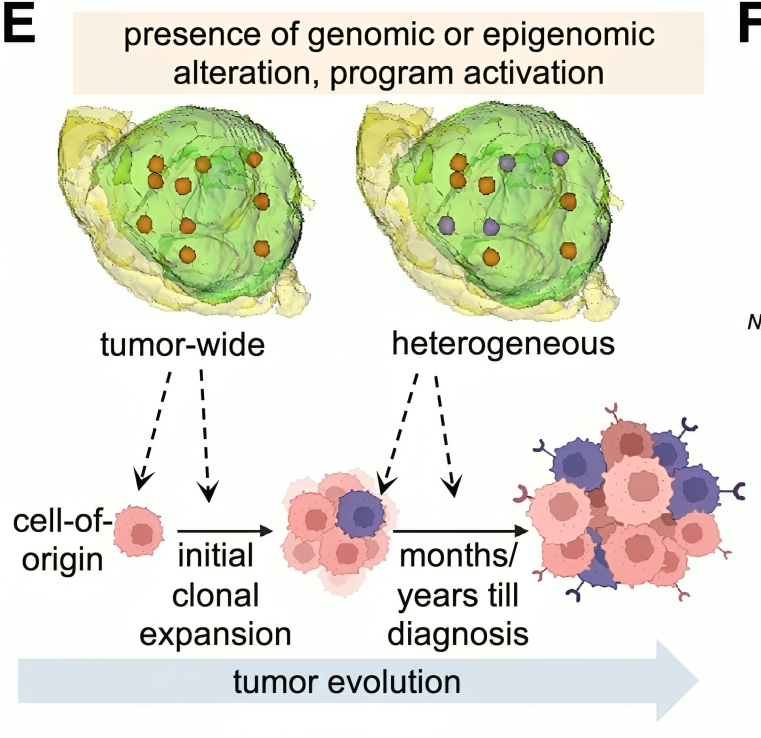
We identified many tumor-wide therapeutic targets that may circumvent heterogeneity-related failures. We also created an online platform for users to explore our 3D spatial map of GBM and interrogate heterogeneity of user-selected genes and programs.
hank you to all our amazing collaborators on this project including Feng Yue from Northwestern Feinberg School of Medicine, Marco Gallo and Ana Nikolic, Nicholas Stevers, Darwin Kwok and many more!
Also very excited to see our work featured in Cell Leading Edge by the The Winkler Lab! Thank you for summarizing our work and highlighting its importance so beautifully!”
Source: Radhika Mathur/X


