Jonathan Keats shared a post on X:
“12 Days of CoMMpass – Day 3: Today we will focus on a common question about how many RNA defined subtypes does the Multiple Myeloma RF CoMMpass study actually identify? Unfortunately, it is a bit of a loaded question, the figures and tables show 12 but in fact it is only 11.
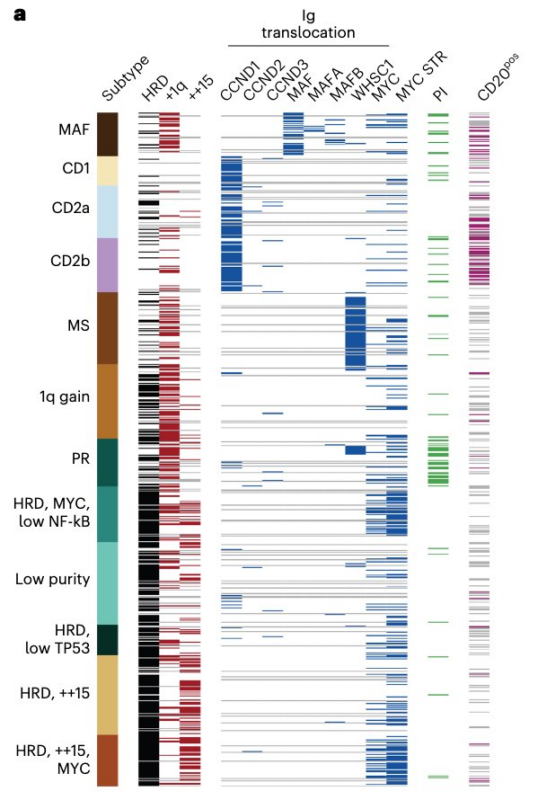
So why the nuance? It all comes down to a ‘subtype’ we call ‘Low Purity’, which matches well with the Myeloid subtype defined by Broijl Annemiek and was pre-removed in Zhan et al, ‘we eliminated samples with high degree of contamination’.
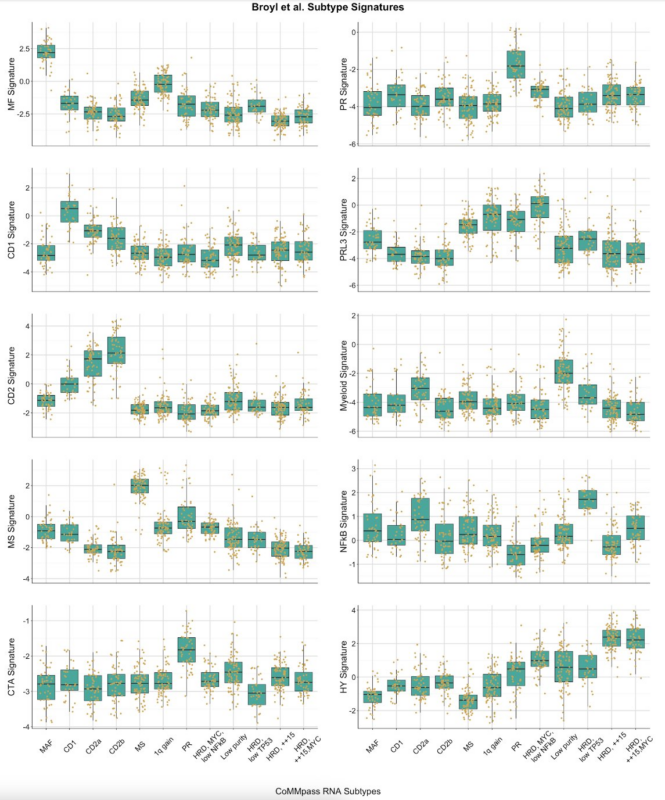
In CoMMpass, we only profiled samples that were >80% tumor content, average 94%, using a slide based kappa/lambda assay. Though in retrospect we likely need different cutoffs for kappa and lambda patients given the different background levels in normal plasma cells.
If you follow the assumption that normal B-cells/plasma cells will be 2/3 kappa and 1/3 lambda expressing, and if an assay like our slide based assay only observed B lineage cells, then 80% lambda can indicate the sample is only 70% tumor while 80% kappa might only mean 40% tumor.
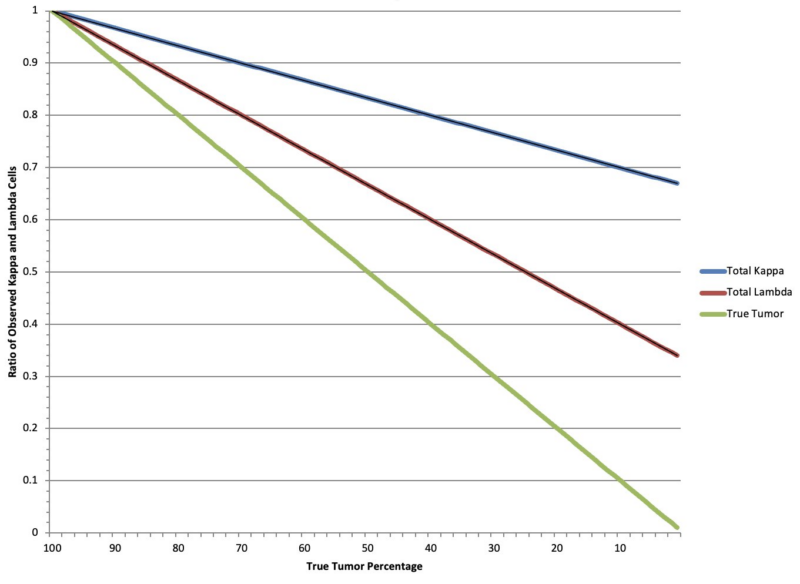
So why do we say this is a low purity group of samples and notice I don’t say subtype? Because we could clearly show there was a correlation with a non B-cell contamination index (RNAseq), a purity estimation (Copy number), and lower somatic mutation allele frequencies.
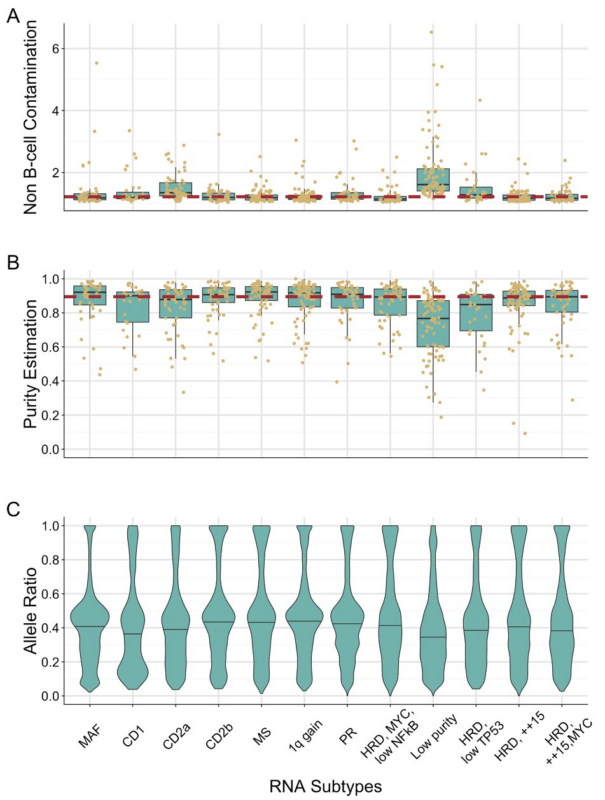
Why did we not filter these samples out like Zhan et al? Largely because with the clustering approach we used when we removed these samples and reclustered it just pulled the next most contaminated into this group. Also, the data indicates the majority of the cell mass is tumor.
Beyond the evidence for low level contamination, we also saw patients with 3 or more timepoints bounce from or into the ‘Low Purity’ group and then back to their disease related subtype.
So it appears this is a feature of the highly dynamic nature of gene expression, the highly divergent gene expression programs in different cell types (contaminating non B-cells) and the variable ability to purify plasma cells for bulk analysis.
The long-term question is what do you do with a clinical test? Inevitably some samples will have this contamination issue, should you want to report subtypes what do you do? I don’t have a good answer but I’d want both DNA and RNA data. Report sample purity as a metric not group.”
Source: Jonathan Keats/X
More posts by Jonathan Keats on oncodaily.com
Jonathan Keats is an assistant professor at Translational Genomics Reseach Institute (TGen). He specialises in Cancer Genetics, Genome Sequencing, and CGH. Previously, he was a research fellow at Mayo Clinic.


