Francesco Maura, Assistant Professor at Sylvester Comprehensive Cancer Center, shared a post on X:
“New paper out in today Nature Communications! With Mayo Clinic, University of Miami, Sylvester Comprehensive Cancer Center, Leif Bergsagel, Marta Chesi, Coffey D we integrated Whole-Genome Sequencing small conditional RNA to show how Vk*MYC Mouse genomic evolution mimics the one observed in human myeloma.
This has been an amazing experience where, thanks to Marta Chesi and Leif Bergsagel. I learned so much about myeloma biology and translational research. Very grateful for the long Zoom meetings and discussions.
Overall we found several interesting data that revealed how Vk*MYC mouse closely mimics multiple myeloma genomic life history.
Several models have been developed to recapitulate the MM evolution and response to therapies. Leif Bergsagel and Marta Chesi developed 15 years ago the Vk*MYC mouse model that after 70-80 weeks spontaneously develops a progressive expansion of monoclonal PC in the BM.
This long evolution is likely driven by the spontaneous selection of additional genomic drivers, beside MYC dysregulation. However, it is largely unknown how well the Vk*MYC MM resembles the genomic landscape and life history of human MM. and this is where our story begun.
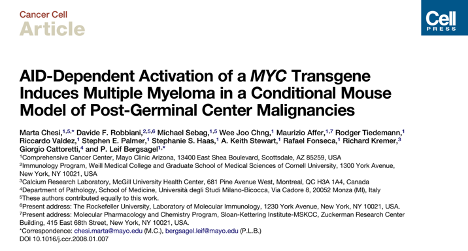
To answer this question, we interrogated the genomic and/or transcriptomic landscape of 119 Vk*MYC MM, including de novo MM, transplantable lines and tumors capable of growing in vitro, to capture the mutations spontaneously selected during disease progression.
Interrogating WGS (n =41) and WES (n = 27), we identified 11 driver genes under positive selection, six of which are related to known driver genes in MM (Dusp2, Trp53, Nfkbia, Pim1, Tent5c/Fam46c, H1f4/Hist1h1e).
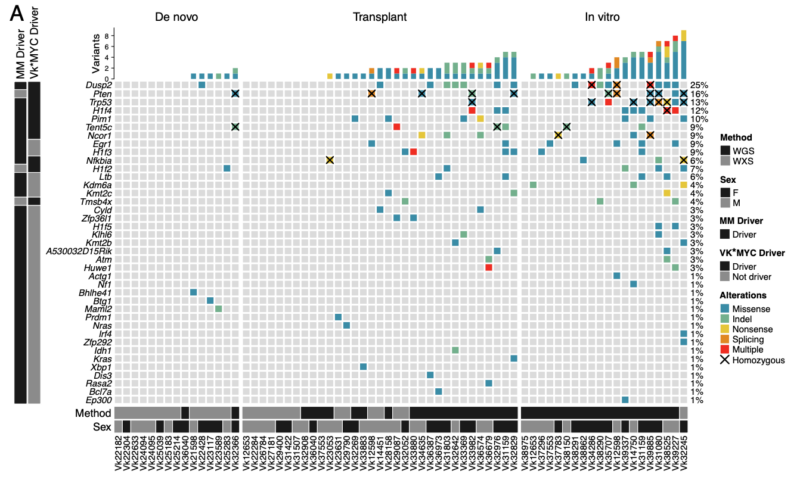
Additional non-synonymous SNVs were found in known human MM driver genes such as Nras, Kras, Cyld, Sp140, Rasa2,and Dis3. Interestingly Tp53 was significantly enriched in In vitro Vk*MYC MM in line with the almost universal p53 inactivation reported in human MM cell lines.
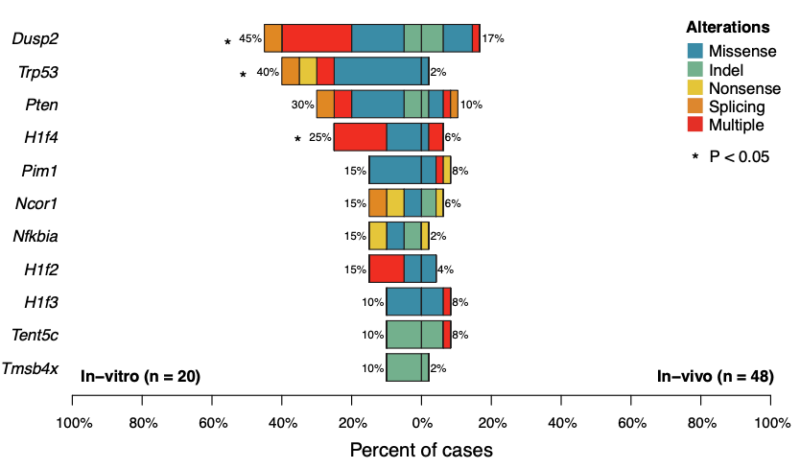
Similarly to human MM, we observed the spontaneous acquisition of several large and focal aneuploidies including several events known to be frequent in human MM such as: gain on chromosome 3 (syngeneic with 1q in human), Pou2af1 gain, Rb1, Ncor1, and Cdkn2a/b loss.
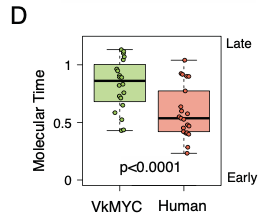
Large duplications were observed in 77% of cases. Using the molecular time approach, we demonstrated that 26% Vk*MYC MM had multiple co-occurring trisomies, similar to what was observed in human MM. In contrast to humans, these gains tended to occur later in time.
Similar to human MM we found evidence of spontaneously acquired chromothripsis in 13% of mice. Complex and single SV events involved several known MM genes including Cdkn2a/b, Kdm6a, Tnfrsf13b, Rb1, Ncor1 and Mcl1.
As an additional similarity between humans and Vk*MYC MM, translocations involving the IGH/IGK/IGL were detected in 8 Vk*MYC MM (15.4%). Some of these events partnered with known oncogenes promoting their overexpression (e.g., Map3k14, Irf4, Pou2af1, and Mcl1)
In addition to these SV, Leif Bergsagel found evidence of LTR insertions resulting in gain of function of Map3k14 and Il6, and gene expression disruption of Ncor1.
Combining SV, SNV and CNV, we noted the convergence of acquired mutations on pathways activating NFkB (38%), RAS/mTORC1 (27%), cell cycle (48%) and chromatin modifiers (67%) as observed in human MM.
It is important to highlight the fact that all these events were spontaneously acquired over 12-18 months, recapitulating the MM genomic evolution and subclonal competition.
We observed the same mutational signatures observed in humans, including SBS84 (AID) and SBS9 (poly-eta in the germinal center), in line with the post-germinal center origin of MM. Including APOBEC !!!! see below and example of a mouse with clear SBS2.
APOBEC is one of the most prevalent genomic defining events in human MM, with strong prognostic implications. APOBEC was confirmed in ~50% of the mice. As in human MM, APOBEC contribution tends to increase from clonal to subclonal variants, suggesting a late role.
Bulk and scRNA data showed that Apobec3 was expressed by both the tumor cells and by normal B-cell and normal plasma cells. In contrast, Apobec1 expression was mostly restricted to the tumor plasma cells, suggesting a potential role in promoting APOBEC mutagenesis.
Most of the APOBEC modelling literature has been using models in which APOBEC is already and/or constitutively active therefore faling to capture the spontaneous APOBEC activation seen in solid and hematological cancer (i.e., MM and lymphomas).
Finally, this study highlights the importance and advantage of applying comprehensive genomic investigations in mouse models in which genomic events are spontaneously acquired over time, and stand as potential models for other tumors.
Thanks to all the collaborators David Coffey Caleb, C. Ola Landgren, Gareth Morgan, Bachisio Ziccheddu and many others!! Again huge thanks to Marta and Leif Bergsagel for this amazing collaboration (and the ones that will follow)!!”
Read further.
Source: Francesco Maura/X


