Biagio Ricciuti shared on X:
“Sharing our latest effort out in Clinical Cancer Research! This multicenter study identified a KEAPness signature that outperforms deleterious KEAP1 mutations in predicting ICI efficacy in NSCLC. A step forward in understanding transcriptional phenocopies of KEAP1 mutations.
Authors: Stefano Scalera, Biagio Ricciuti, Daniele Marinelli, Marco Mazzotta, Laura Cipriani, Giulia Bon, Giulia Schiavoni, Irene Terrenato, Alessandro Di Federico, Joao V. Alessi, Maurizio Fanciulli, Ludovica Ciuffreda, Francesca De Nicola, Frauke Goeman, Giulio Caravagna, Daniele Santini, Ruggero De Maria, Federico Cappuzzo, Gennaro Ciliberto, Mariam Jamal-Hanjani, Mark M. Awad, Nicholas McGranahan, Marcello Maugeri-Saccà.

KEAP1 mutations are frequent drivers of primary resistance to ICI in NSCLC. However, these mutations only identify a fraction of non-responders to immunotherapies.
We hypothesized that a fraction of KEAP1 wild-type tumors recapitulates the transcriptional footprint of KEAP1 mutations, and that this KEAPness phenotype outperforms mutation-based models in predicting ICI efficacy.
We used differential gene expression and unsupervised hierarchical clustering between tumors with and without KEAP1-NRF2 pathway mutations and identified a KEAPness signature. This signature was present across ~ 8,000 KEAP1 wild-type tumors spanning 17 cancer types.
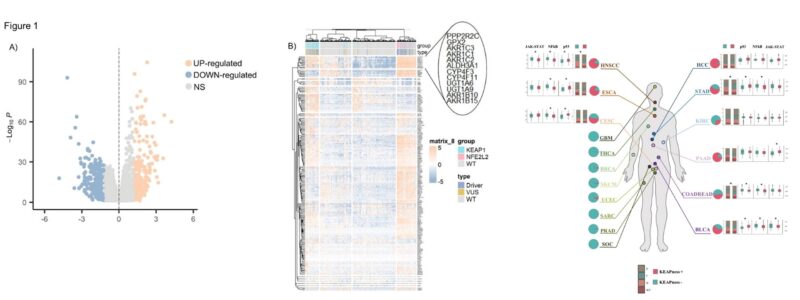
Similarly to KEAP1 mutations, this KEAPness signature was associated with an immune-excluded microenvironment, decreased MHC-I/II expression and decreased T cells density.
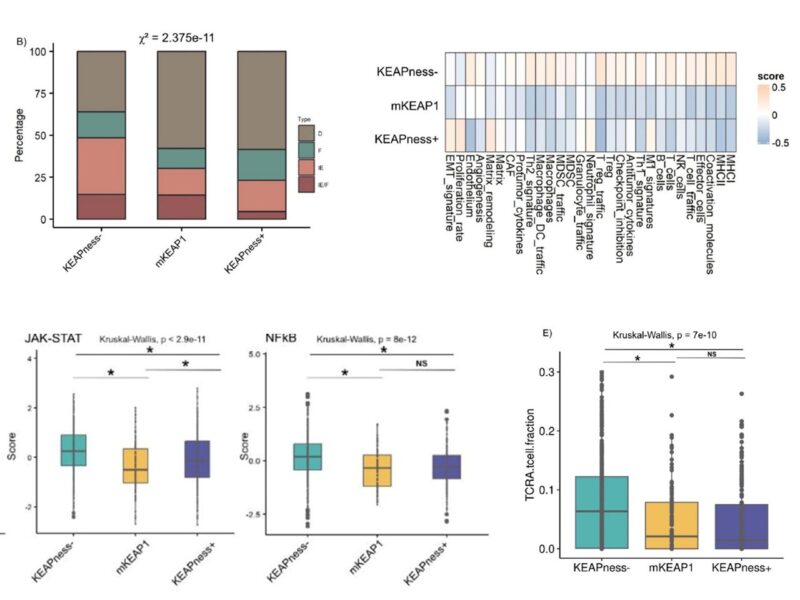
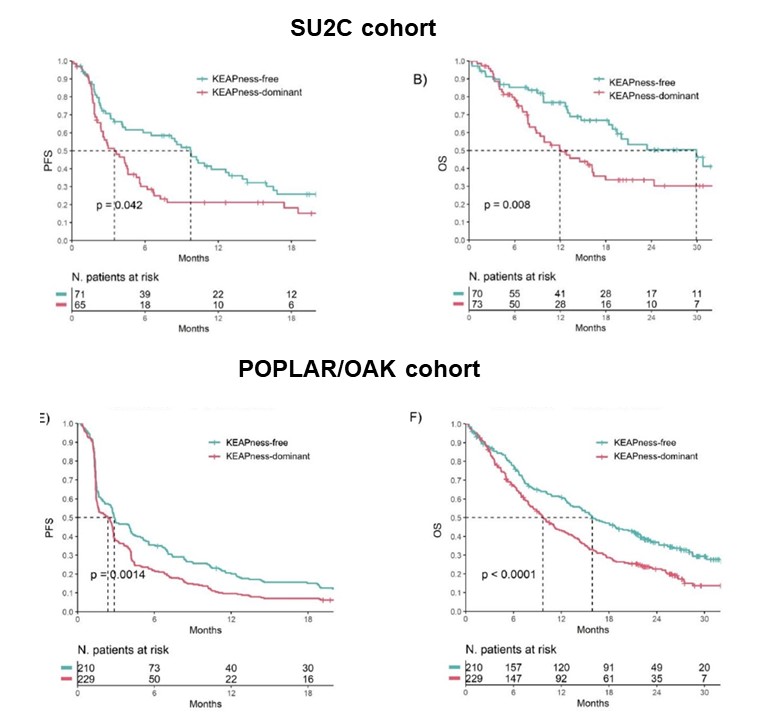
Because of this immune-excluded microenvironment, patients with tumors harboring KEAPness signature experienced inferior PFS and OS with immunotherapy compared to patients w/ KEAPness negative tumors. This was confirmed in 2 independent cohorts.
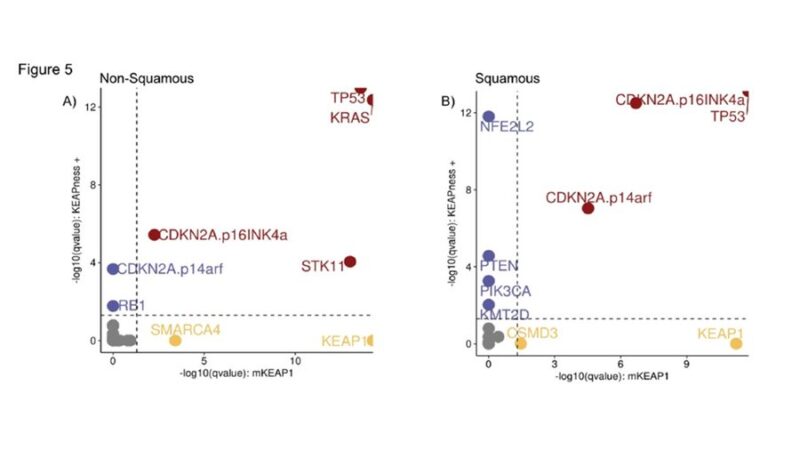
Genetic selection and evolutionary trajectories analysis in the TRACERx 421 cohort revealed that KEAPness+ and KEAP1-mutant Nsq-NSCLC shared positive selection for TP53/KRAS/STK11. CDKN2A (p14arf) was identified as a private driver in KEAPness+ but not KEAP1-mutant samples.
In summary, here we reported that transcriptional phenocopies of KEAP1 mutations are common in NSCLC and are associated with immune exclusion, and resistance to ICI, independently from the presence of KEAP1 mutations.
These results suggest that integrating genomic and transcriptomic profiling can help: 1) identify patients who may not respond to PD-(L)1 monotherapy with more accuracy and 2) redirect them to the most appropriate immunotherapy approach.”
Source: Biagio Ricciuti/X
Biagio Ricciuti is a thoracic oncologist at the Dana-Farber Cancer Institute and an Assistant Professor of Medicine at Harvard Medical School.
His research interests are various aspects of lung cancer, including novel therapeutics, biomarker identification, and predictive modeling for patient outcomes. Besides being actively engaged in research, Dr. Ricciuti is involved in clinical trials and patient care.


