Jonathan Keats shared on X:
“12 Days of CoMMpass – Day 2: The intention of these daily threads is to pay homage to the 12 years of effort in the Multiple Myeloma Research Foundation CoMMpass study. This study accrued myeloma patients over 4 years starting July 2011 through June 2015 and followed patients for 8 years, ending June 2.
The study was formulated after the seismic shift in our treatment paradigms with immunomodulatory drugs and proteasome inhibitors becoming the foundation for frontline therapy. Thankfully, the available therapies have improved even more during the CoMMpass study.
Today we will focus on the RNA subtype call MS that is associated with t(4;14) positive multiple myeloma with 92.5% of patients having a t(4;14) detected by whole genome sequencing. This event causes overexpression of NSD2, formerly WHSC1, and known in Multiple Myeloma as MMSET.
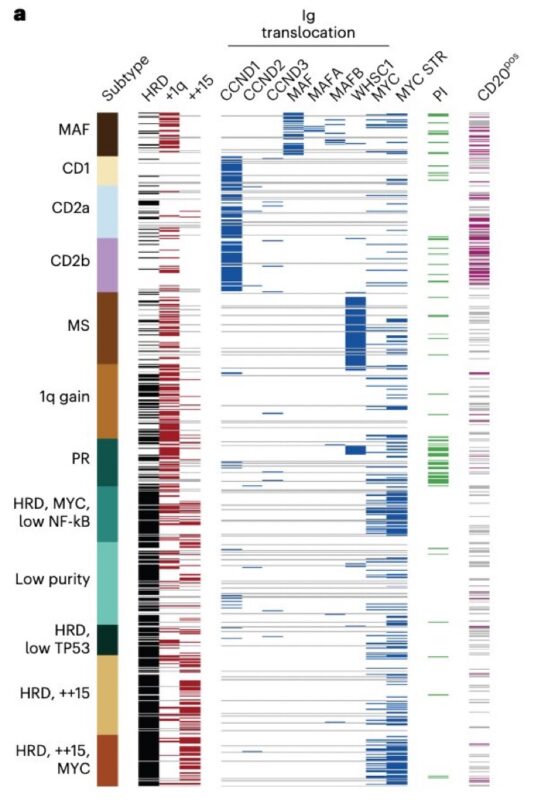
Question: What is going on in the 5 patients without t(4;14) detected? Thanks to whole genome sequencing we can see one is a rare kappa locus translocation resulting in NSD2/WHSC1/MMSET overexpression by the kappa enhancer.
Two others had translocations that created fusion genes with FUT8::NSD2 and CXCR4::NSD2. Due to the high expression of these genes in plasma cells the fusion transcripts, like the IGH::NSD2 fusions created by t(4;14) are highly expressed.

These events tell us the MS subtype is driven by NSD2 overexpression not t(4;14). If we put t(4;14) into a high-risk category should we not also put these three edge case patients into a high risk group as well? TGen we will do it with rapid WGS in the TGen clinical lab.
So, what about the 2 patients I have not explain? These reflect the limitations of the shallow long-insert WGS used in CoMMpass. Which we developed in 2010 as a cost effective way to detect genome wide copy number and structural events with 6-8x genome coverage.
In one patient there is a deletion of 4p16, commonly seen in single fusion FISH case lacking FGFR3 overexpression and a single informative read in the common breakpoint locus in intron 1 of LETM1 suggesting a t(4;14) does exist.
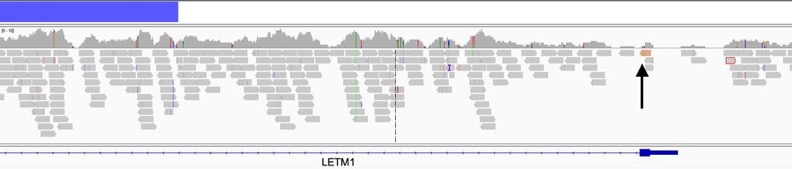
In the last patient we see a similar event with a 4p16 deletion, but in this case there are 3 informative reads, one less than the automated calling requirement. Importantly, for future NGS testing this common breakpoint at NSD1 exon 1 is prone to drop-out due to high GC content.

Takeaway: If you think t(4;14) is important, you need to realize what is really important is NSD2 overexpression, and there is more than one way this occurs in myeloma. We need better tests to detect all of these events, because if you are the edge case patient it matters!”
Source: Jonathan Keats/X
Jonathan Keats is an assistant professor at Translational Genomics Reseach Institute (TGen). He specialises in Cancer Genetics, Genome Sequencing, and CGH. Previously, he was a research fellow at Mayo Clinic.


