EWSR1 Gene Rearrangements in Cancer: Mechanisms and Clinical Implications
EWSR1 gene is among the most studied chromosomal abnormalities, whose rearrangements, such as EWSR1::ATF1 fusions, alter transcriptional regulation and contribute to oncogenesis.
The EWSR1 gene, frequently rearranged in diverse cancers, exemplifies the impact of chromosomal damage on transcriptional regulation and tumorigenesis.
These changes have significant implications for diagnostics and therapy, particularly in developing tumor-agnostic approaches.
Study led by Julia Raffaella Bianco and colleagues provided an in-depth analysis of the EWSR1::ATF1 fusion, its mechanisms, and its implications in oncogenesis
EWSR1::ATF1 Translocation: A Common Tumor Driver of Distinct Human Neoplasms
Authors: Julia Raffaella Bianco, YiJing Li, Agota Petranyi, Zsolt Fabian

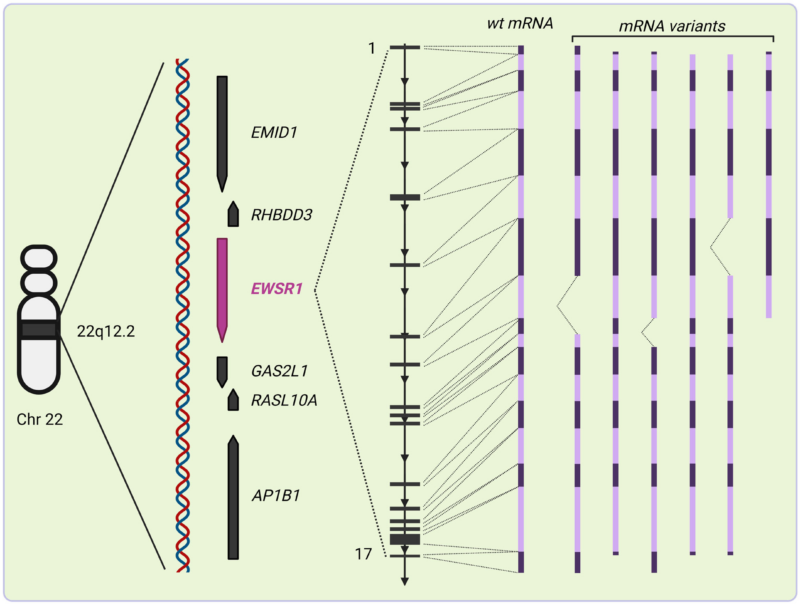
The EWSR1 gene, located on chromosome 22q12.2, encodes a 656-amino acid protein within the conserved FET family, which includes FUS and TAF15. The EWSR1 promoter region lacks a TATA box, supports multiple transcription start sites, and is regulated by WNT/β-catenin signaling.
Alternative splicing generates 22 transcript variants, seven of which are non-coding. These variants influence tumor behavior and progression, with some facilitating oncogenesis through protein alterations.
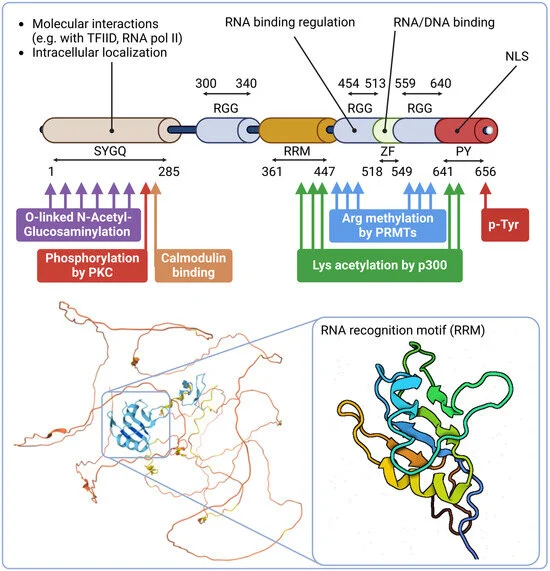
The EWSR1 transcript spans 40 kb with 17 exons, frequently disrupted in tumors like Ewing sarcoma due to breakpoints in introns 6–8. The N-terminal LCD domain of EWSR1 mediates transcriptional activation and phase separation, critical in oncogenesis. The LCD domain is regulated by post-translational modifications like O-linked glycosylation, which alters localization and interaction dynamics, and serine/threonine phosphorylation.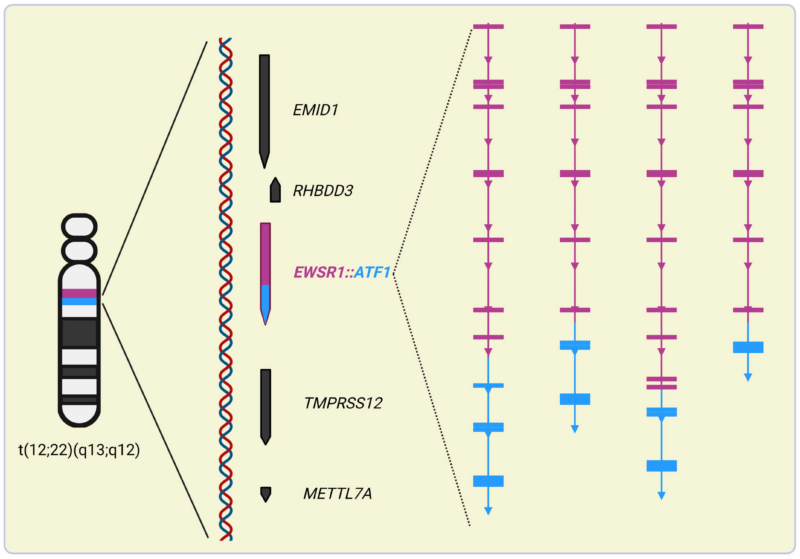
The RNA-binding domain (RBD) in the mid-region of EWSR1 includes an RNA recognition motif and RGG boxes that facilitate RNA and DNA interactions, regulating transcription, splicing, and miRNA biogenesis.
Chromosomal translocations involving EWSR1 are common in soft-tissue tumors, fusing its N-terminal domain with DNA-binding domains of partners like ATF1.
These chimeric proteins disrupt normal gene expression and signaling, driving tumorigenesis. Intronic Alu elements in EWSR1 are implicated in these rearrangements, with population-specific allele frequencies correlating with cancer incidence.
Overall, EWSR1 plays diverse roles in transcription, DNA repair, and RNA metabolism, with its dysregulation and fusion events significantly contributing to cancer pathogenesis.
ATF1: Structure, Function, and Oncogenic Implications
The ATF1 gene, located on chromosome 12q13, encodes a 271-amino acid protein that belongs to the bZIP (basic leucine zipper) family of transcription factors. It is flanked by the DIP2B and TMPRSS12 genes and spans approximately 57 kb.
The gene structure consists of seven exons separated by six introns, with exon 2 containing the initiation codon. The gene’s intronic regions feature repetitive sequences, including Alu elements, which may contribute to genomic instability.
Translation of ATF1 mRNA results in three protein-coding isoforms (ATF1-201, -204, and -205), while two additional transcripts undergo nonsense-mediated mRNA decay. Regulation of ATF1 expression is mediated through interactions between its 3′ UTR and microRNAs such as miR-34c, which represses ATF1 at the post-transcriptional level.
Protein Structure and Functional Domains
The ATF1 protein contains several domains critical for its function:
1. Basic-Leucine Zipper (bZIP): Located in the C-terminus and encoded by exons 6 and 7, the bZIP domain enables DNA binding and dimerization with other bZIP proteins such as CREB and AP-1. The domain binds cAMP-responsive promoter elements (CRE) and is essential for transcriptional regulation.
2. N-Terminal Region (NTR): Unlike its homolog CREB, ATF1’s NTR lacks certain transcriptionally active regions but can be phosphorylated at serine residues, which destabilizes DNA binding.
3. Phosphorylated Kinase-Inducible Domain (pKID): Encoded by exon 3, pKID includes phosphorylation sites for multiple kinases, such as PKA and CDK3, which modulate transcriptional activity.
Role in Cellular Processes
ATF1 regulates genes involved in cell proliferation, differentiation, survival, and immune response. It binds to over 15,000 genomic sites, influencing genes in pathways such as apoptosis (BCL2/BAX ratio) and T-cell activation. ATF1 also interacts with BRCA1, enhancing its role in the DNA damage response and transcriptional activation.
Oncogenic Role
Phosphorylation of ATF1 at specific sites (e.g., threonine 184) stabilizes its expression and correlates with metastasis in certain cancers, including gastric carcinoma and melanoma. Elevated ATF1 expression drives oncogenesis by regulating genes associated with tumor proliferation, survival, and metastasis, such as matrix metalloproteinase-2 (MMP2).
Mechanisms and Clinical Implications
The EWSR1::ATF1 fusion arises from translocations, most commonly involving intron 3 of ATF1 and intron 8 of EWSR1. This fusion produces an oncogenic chimeric protein, with the transcriptional activation domain of EWSR1 fused to the DNA-binding bZIP domain of ATF1.
Functional Characteristics
The EWSR1::ATF1 protein differs from wild-type ATF1 in several ways:
1. Transcriptional Retargeting: The fusion alters binding specificity, leading to increased binding at distal genomic sites and activation of novel target genes.
2. Enhanced Transcriptional Activity: The EWSR1 domain recruits additional co-factors and transcriptional machinery, amplifying gene expression.
3. Altered Signaling Pathways: While PKA signaling is often excluded, CKII-mediated phosphorylation may enhance the fusion protein’s DNA-binding activity.
Tumorigenesis and Tumor Types
The EWSR1::ATF1 fusion is associated with several rare malignancies, including:
– Clear Cell Sarcoma (CCS): A highly aggressive soft-tissue tumor with poor prognosis. CCS often mimics melanoma but can be differentiated by the presence of EWSR1::ATF1.
– Angiomatoid Fibrous Histiocytoma (AFH): A less aggressive tumor commonly found in soft tissues, often presenting as painless masses.
– Intracranial Non-Myxoid AFH (iAFH): A rare pediatric central nervous system tumor with nonspecific clinical symptoms.
– Gastrointestinal Neuroectodermal Tumors (GNETs): Aggressive tumors with neural differentiation markers.
– Pediatric Malignant Mesothelioma: A subtype distinct from asbestos-induced adult mesothelioma.
Diagnostic and Therapeutic Implications
The identification of EWSR1::ATF1 fusion serves as a diagnostic marker for these malignancies. While traditional therapies often show limited efficacy, emerging treatments, such as histone deacetylase inhibitors, have demonstrated potential in repressing EWSR1::ATF1 activity.
This fusion exemplifies the role of chimeric proteins in driving tumorigenesis and highlights the need for targeted therapeutic strategies.
EWSR1’s role as a biomarker and therapeutic target highlights the need for further research into its molecular mechanisms to advance cancer diagnostics and develop targeted therapies.


