Personalized Immunotherapy in Lung Adenocarcinoma via Single-Cell Profiling of MAFF⁺ Tumors
Lung adenocarcinoma (LUAD) is a biologically complex and clinically heterogeneous malignancy, characterized by multiple oncogenic drivers and a highly immunosuppressive tumor microenvironment (TME). Although targeted therapies and immune checkpoint inhibitors (ICIs) have improved outcomes in subsets of patients, therapeutic resistance and disease progression remain major challenges. Increasing evidence suggests that tumor–immune ecosystem dynamics, particularly those involving myeloid cells, shape disease evolution and therapy response. Using single-cell RNA sequencing (scRNA-seq), this study reveals a distinct LUAD tumor cell population driven by the transcription factor FOS, which orchestrates both tumor-intrinsic and extrinsic immune evasion programs.
Methods Overview
The analysis leveraged publicly available scRNA-seq data (GSE164789) encompassing 31 LUAD samples. Cell clustering identified 14 major cell types, with subsequent refinement of tumor epithelial cells using inferCNV and functional annotations via CellChat (for intercellular communication), Monocle (for trajectory analysis), CytoTRACE (for stemness), and SCENIC (for transcription factor regulatory networks). Functional validation of key findings was performed in LUAD cell lines (A549, H1975) using siRNA knockdown of FOS. A 12-gene MAFF+ Tumor Risk Score (MTRS) prognostic model was constructed to assess patient survival, immune landscape, and drug sensitivity.
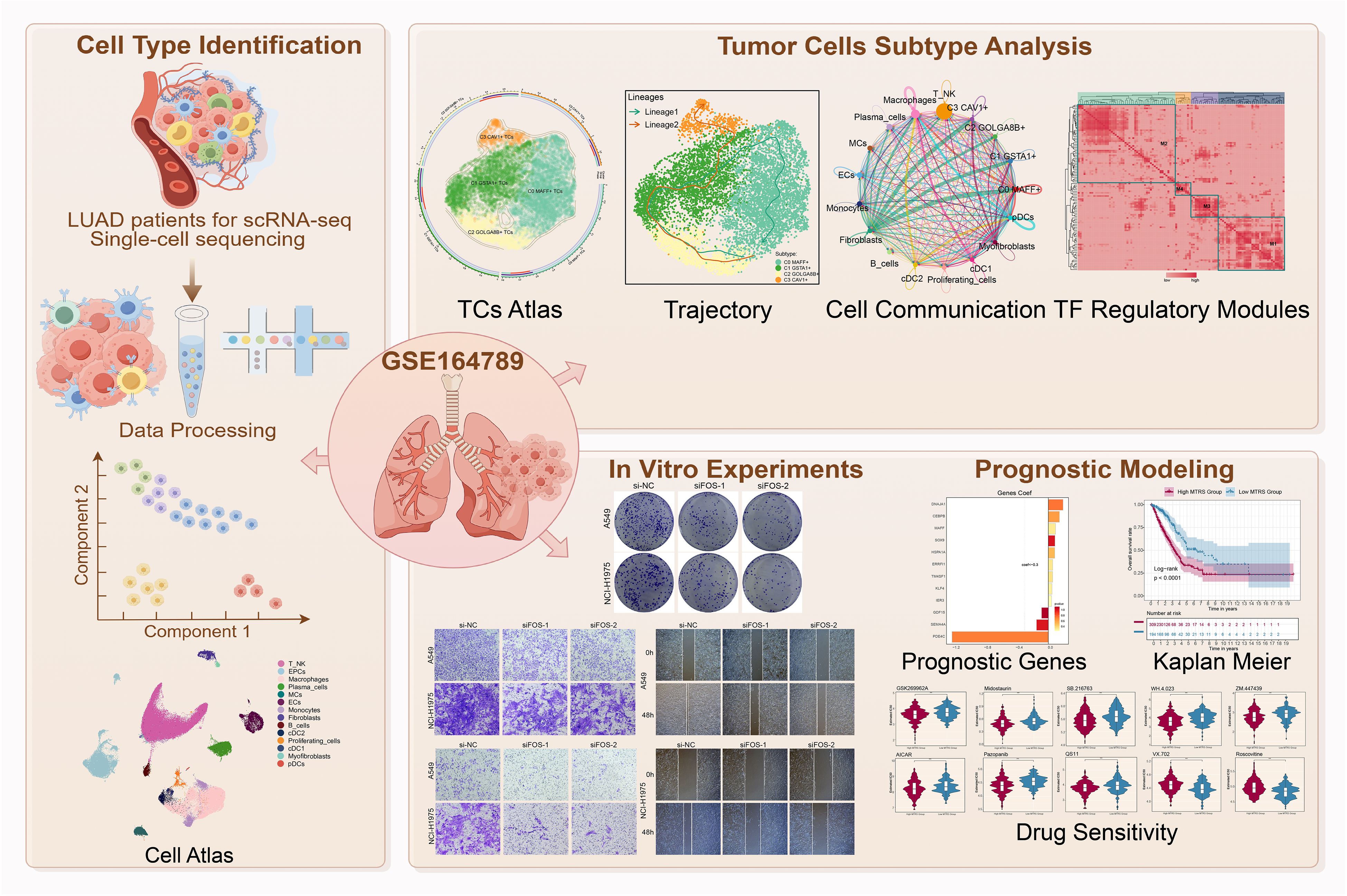
Discovery of the MAFF+ Tumor Subtype (C0)
Among the four tumor subtypes (C0–C3), the C0 subtype emerged as the most aggressive. These tumor cells expressed high levels of MAFF, along with key regulators of lipid metabolism (e.g., SCD, PLIN2) and inflammatory cytokines (CXCL1, CXCL3). C0 cells demonstrated:
- High stemness potential (CytoTRACE score >0.8),
- Low differentiation status (based on trajectory analysis),
- Enrichment in lipid metabolic pathways, associated with tumor progression and immune escape,
- Strong potential for immune–stromal interaction, particularly with macrophages and fibroblasts.
These features suggest that C0 cells represent a stem-like, pro-metastatic tumor phenotype with elevated immune evasion capabilities.
Immune Evasion via Macrophage Polarization
CellChat analysis identified MIF–(CD74+CD44) as a dominant ligand–receptor pair mediating communication between C0 tumor cells and macrophages. This signaling axis is known to:
- Promote macrophage M2 polarization,
- Enhance angiogenesis and tissue remodeling,
- Suppress antigen presentation and T cell activation.
The study confirmed that increased CD44+CD74+ TAMs were present in C0-enriched tumors, supporting a model in which MAFF+ tumor cells actively reprogram the innate immune microenvironment to facilitate immune suppression and tumor progression.
FOS as a Master Regulator of Tumor Aggressiveness
Regulatory network analysis using the SCENIC pipeline identified a distinct transcriptional signature in the C0 tumor cell population, with this transcription factor emerging as one of the top-ranking regulators uniquely enriched in this poorly differentiated, plastic cell state. Functionally, this factor is a key component of the AP-1 transcriptional complex, partnering with proteins such as JUN and ATF3 to orchestrate a wide array of cellular programs central to tumor aggressiveness.
Among its critical regulatory roles, this transcription factor is known to modulate cell proliferation and invasion, directly influencing tumor growth and metastatic potential. It is also a potent driver of the epithelial–mesenchymal transition (EMT), a process by which epithelial cancer cells acquire mesenchymal features that facilitate dissemination and therapeutic resistance.
Importantly, the transcription factor has been implicated in the transcriptional activation of genes involved in lipid biosynthesis, aligning with the observed enrichment of metabolic pathways in C0 cells. This metabolic rewiring may endow these cells with enhanced plasticity and survival capacity under stress conditions within the tumor microenvironment. Moreover, it plays a prominent role in inflammatory signaling cascades, including IL6/JAK/STAT3 and TNF/NF-κBpathways—both of which are known to contribute to immunosuppressive niche formation and resistance to immune checkpoint blockade.
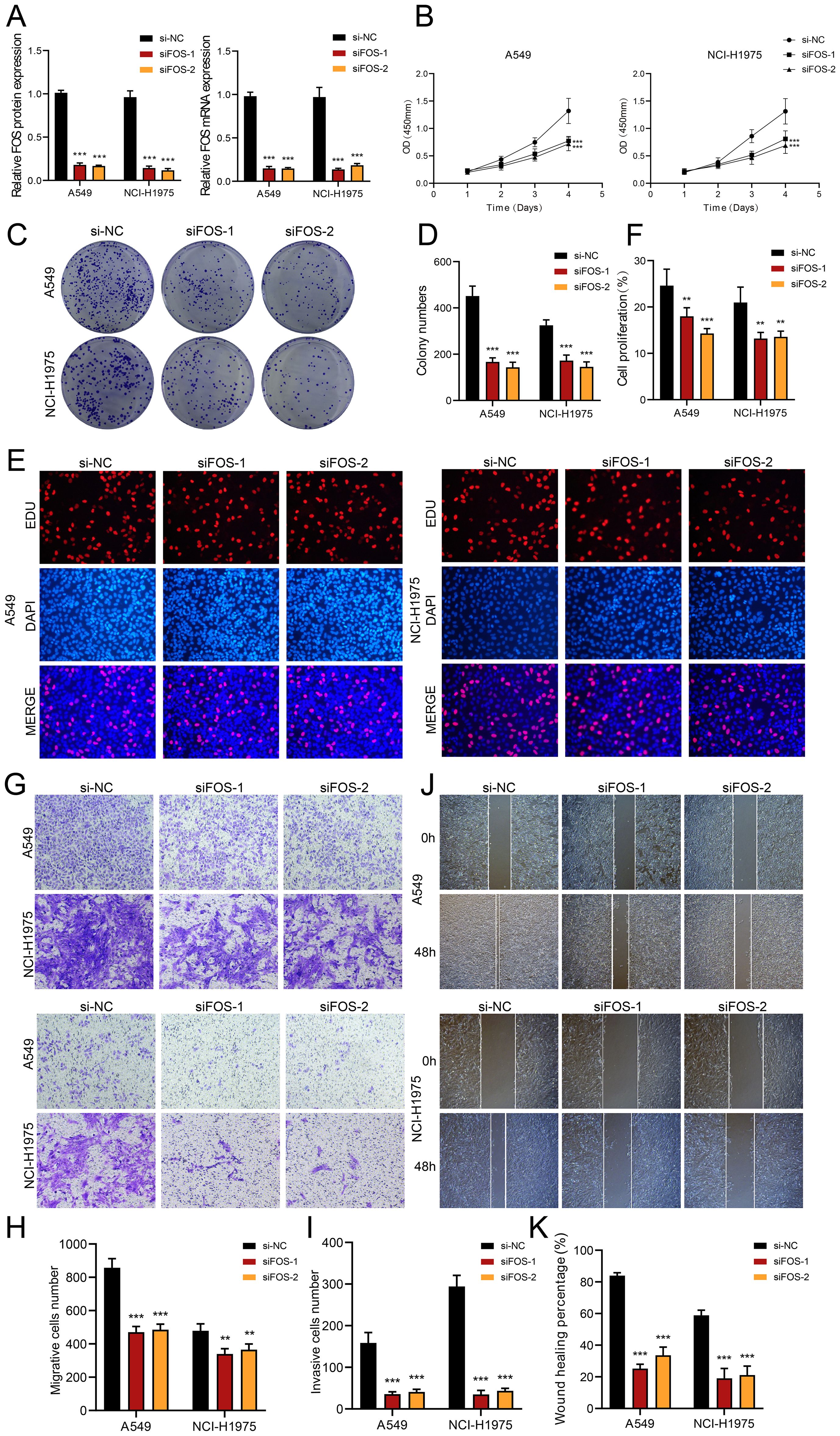
Functional Validation of FOS Oncogenicity
In vitro knockdown experiments in A549 and H1975 cells showed that FOS silencing significantly reduced:
- Proliferation (colony formation),
- Migration and invasion (transwell assays),
- Expression of EMT and lipid metabolism markers, such as SNAI1, SCD, and PLIN2.
These results establish FOS as a tumor-intrinsic oncogene and a regulator of pro-tumorigenic cell states. Importantly, FOS also emerges as a bridge between metabolic reprogramming and immune modulation, supporting its candidacy as a multi-dimensional therapeutic target.
Single-cell transcriptomic profiling revealed a striking enrichment of a distinct transcriptional program within the C0 tumor cell population, characterized by high plasticity and low differentiation status. Central to this program was the selective overexpression of a specific transcription factor (later identified as FOS), which was notably absent in more differentiated tumor subtypes, including alveolar-like and cycling epithelial cells. This selective expression pattern suggests a lineage-restricted regulatory role tightly linked to tumor cell stemness.
Further analysis uncovered a positive correlation between this transcription factor and genes involved in lipid metabolism, implicating the existence of a metabolic–transcriptional axis that may endow these tumor cells with enhanced adaptability and survival under metabolic stress. The upregulation of lipid metabolic pathways within the same cell state supports the notion that this transcription factor may coordinate both transcriptional plasticity and bioenergetic flexibility in the tumor microenvironment.
Importantly, elevated expression of this transcriptional regulator also showed a strong association with increased macrophage infiltration, suggesting an indirect yet potent role in shaping the tumor immune landscape. This connection indicates that beyond intrinsic effects on tumor cell behavior, the transcription factor may influence the recruitment or polarization of myeloid cells, contributing to an immunosuppressive microenvironment that facilitates disease progression.
Functional Validation of FOS Oncogenicity
To validate the functional relevance of this transcription factor in EGFR-mutated lung adenocarcinoma, in vitro silencing experiments were conducted using siRNA in two representative NSCLC cell lines: A549 and H1975. Suppression of expression led to a marked reduction in cellular proliferation, as evidenced by a significant decrease in colony formation. Additionally, transwell migration and invasion assays revealed that knockdown substantially impaired the cells’ ability to migrate and invade—hallmarks of aggressive tumor behavior.
At the molecular level, knockdown resulted in a concurrent downregulation of key markers associated with epithelial–mesenchymal transition (EMT) and lipid metabolism, including SNAI1 (a transcriptional driver of EMT), SCD (a lipogenic enzyme), and PLIN2 (a lipid droplet-associated protein). These findings support the notion that this transcription factor promotes a pro-tumorigenic cell state, integrating proliferative, invasive, and metabolic programs.
Importantly, this regulatory role appears to extend beyond intrinsic tumor behavior. By coordinating both metabolic reprogramming and immune-modulatory signals, the factor functions as a central node linking tumor cell plasticity to microenvironmental interactions. These data establish it as a multi-dimensional oncogenic driver and nominate it as a promising therapeutic target in EGFR-mutated NSCLC.
MTRS Model and Therapeutic Implications
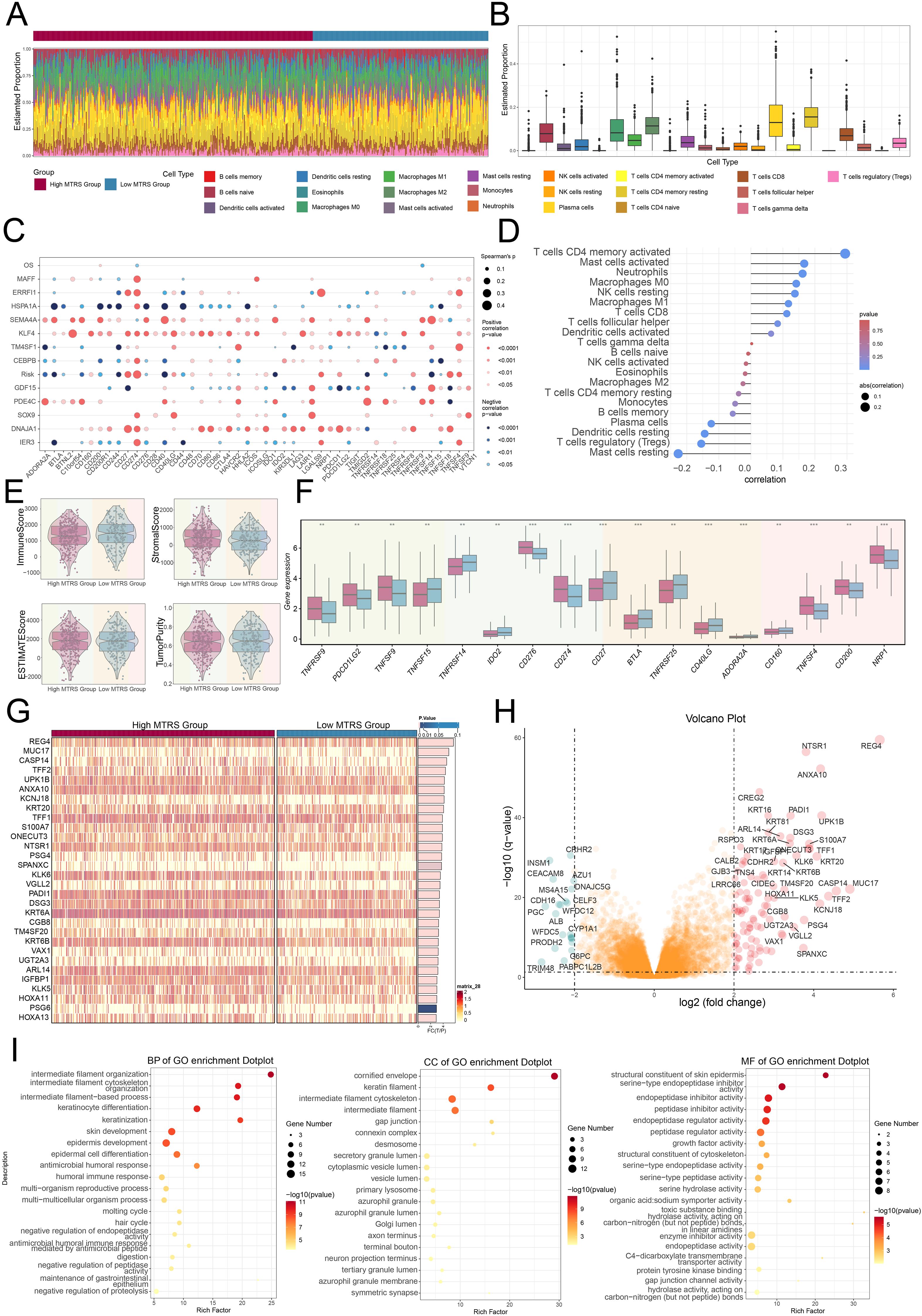
To translate the molecular characteristics of the C0 subtype into clinical utility, the investigators developed a 12-gene Tumor Risk Score model, termed the MAFF⁺ Tumor Risk Score (MTRS). This gene signature was derived from differentially expressed genes enriched in the C0 cluster and was designed to quantify individual tumor risk. The MTRS effectively stratified patients into high-risk and low-risk groups, with the high-risk cohort demonstrating significantly poorer overall survival. Importantly, this prognostic capability was shown to be independent of clinical stage, underscoring the added value of transcriptomic profiling beyond conventional staging systems.
Using CIBERSORTx immune deconvolution, the high MTRS group was also found to be enriched for macrophage infiltration, aligning with the immunosuppressive and metabolically reprogrammed landscape previously associated with C0 tumors. Furthermore, transcriptomic analysis revealed that immune checkpoint molecules, including PD-L1 and LAG3, were more highly expressed in the high-risk subgroup, suggesting a potential for immune evasion and resistance in this population.
In addition to its prognostic power, the MTRS also revealed therapeutic vulnerabilities. Drug sensitivity predictions identified several compounds with selective efficacy in high-risk tumors, including AICAR (an AMPK activator targeting metabolic stress), Midostaurin (a broad-spectrum kinase inhibitor), and GSK269962A (a ROCK inhibitor affecting cytoskeletal dynamics). These agents hold promise for modulating lipid metabolism, invasive phenotypes, and stromal interactions, potentially offering targeted avenues for intervention in aggressive molecular subtypes.
Future Perspectives
- Clinical trials are warranted to test FOS inhibitors (e.g., T-5224) in LUAD, particularly in patients with high MTRS scores.
- Combination approaches targeting MIF–CD74/CD44 signaling alongside PD-1/PD-L1 blockade may improve efficacy.
- Further exploration of lipid metabolism inhibitors could suppress the stem-like, immune-evasive MAFF+ tumor states.
- Integration of scRNA-seq and bulk transcriptomic signatures into diagnostic workflows may facilitate biomarker-guided treatment.
You Can Read The Full Article Here
Written by Toma Oganezova, MD, Editor-in-Chief of OncoDaily IO
-
Challenging the Status Quo in Colorectal Cancer 2024
December 6-8, 2024
-
ESMO 2024 Congress
September 13-17, 2024
-
ASCO Annual Meeting
May 30 - June 4, 2024
-
Yvonne Award 2024
May 31, 2024
-
OncoThon 2024, Online
Feb. 15, 2024
-
Global Summit on War & Cancer 2023, Online
Dec. 14-16, 2023
