T cell therapies have revolutionized the treatment of hematologic malignancies, offering curative potential in several blood cancers. However, their success in solid tumors has been limited, largely due to persistent challenges inherent to the tumor microenvironment. Among the most pressing barriers are the inadequate infiltration of T cells into tumor sites, the presence of a highly immunosuppressive milieu that dampens immune responses, and the development of T cell exhaustion that compromises functionality over time.
In response to these obstacles, the authors of this study designed a cutting-edge in vivo genome-wide CRISPR knockout screening platform. By utilizing primary human T cells within mouse models bearing solid tumors, they sought to systematically identify genetic modifications capable of enhancing T cell persistence, infiltration, and effector function—ultimately aiming to improve the efficacy of T cell therapies in the context of solid malignancies.
Methodology and Model Development
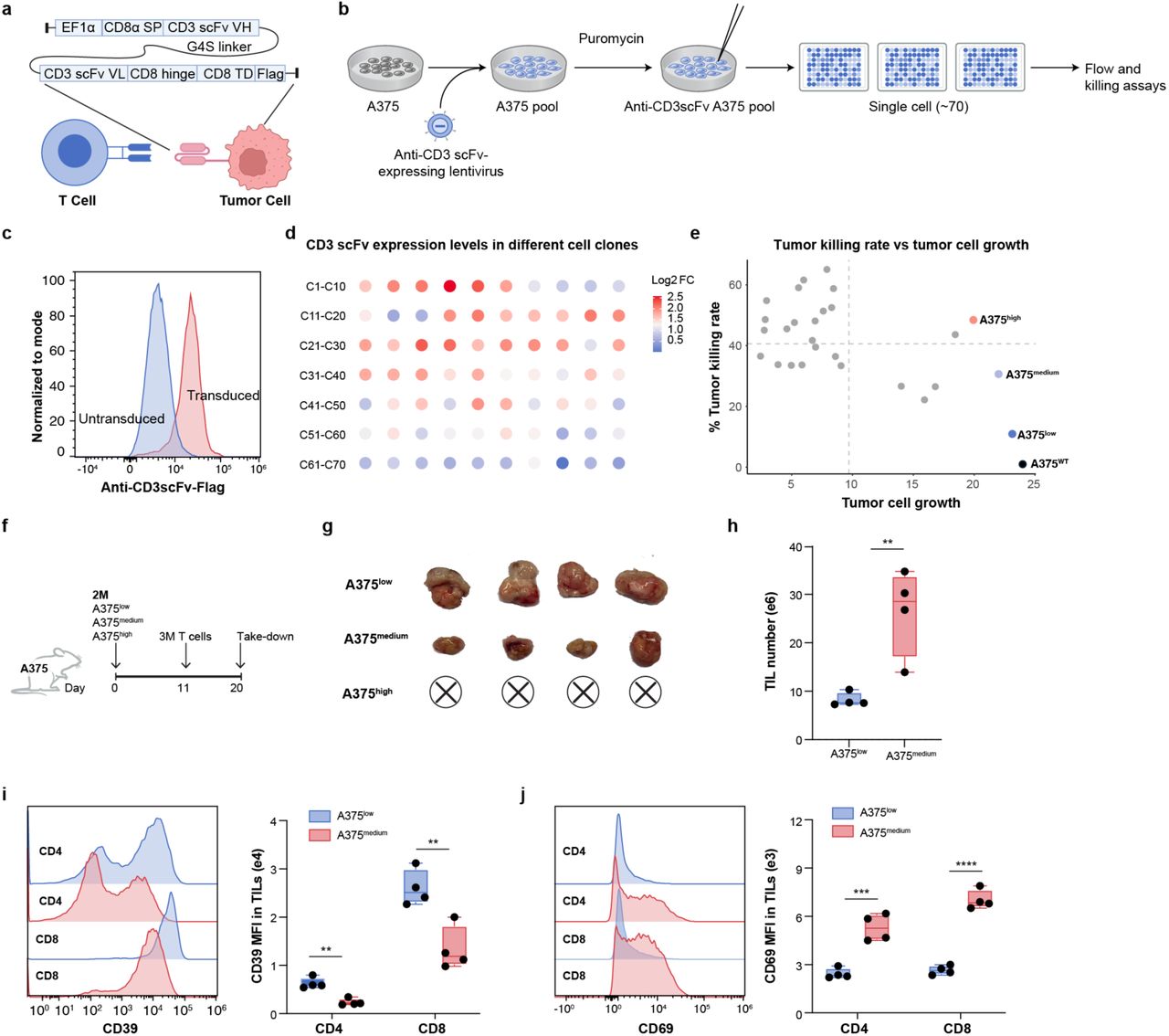
To enable large-scale functional screening of tumor-infiltrating T cells (TILs), the researchers engineered a novel tumor model by introducing an anti-CD3 single-chain variable fragment (scFv) into human A375 melanoma cells. This modification created an artificial immune synapse that actively recruited and engaged T cells within the tumor, dramatically increasing their infiltration. As a result, millions of TILs could be recovered from individual tumors, providing sufficient cell numbers for high-throughput CRISPR screening.
Importantly, the T cells recovered from the tumor microenvironment displayed hallmark features of exhaustion—such as CD39⁺ and PD-1⁺ expression—alongside marked functional impairments compared to their splenic counterparts. This recapitulation of TIL dysfunction observed in patients underscores the model’s clinical relevance and its utility for identifying genetic interventions to restore T cell activity in solid tumors.
Genome-Wide CRISPR Screens Reveal New Targets to Enhance T Cell Immunotherapy in Solid Tumors
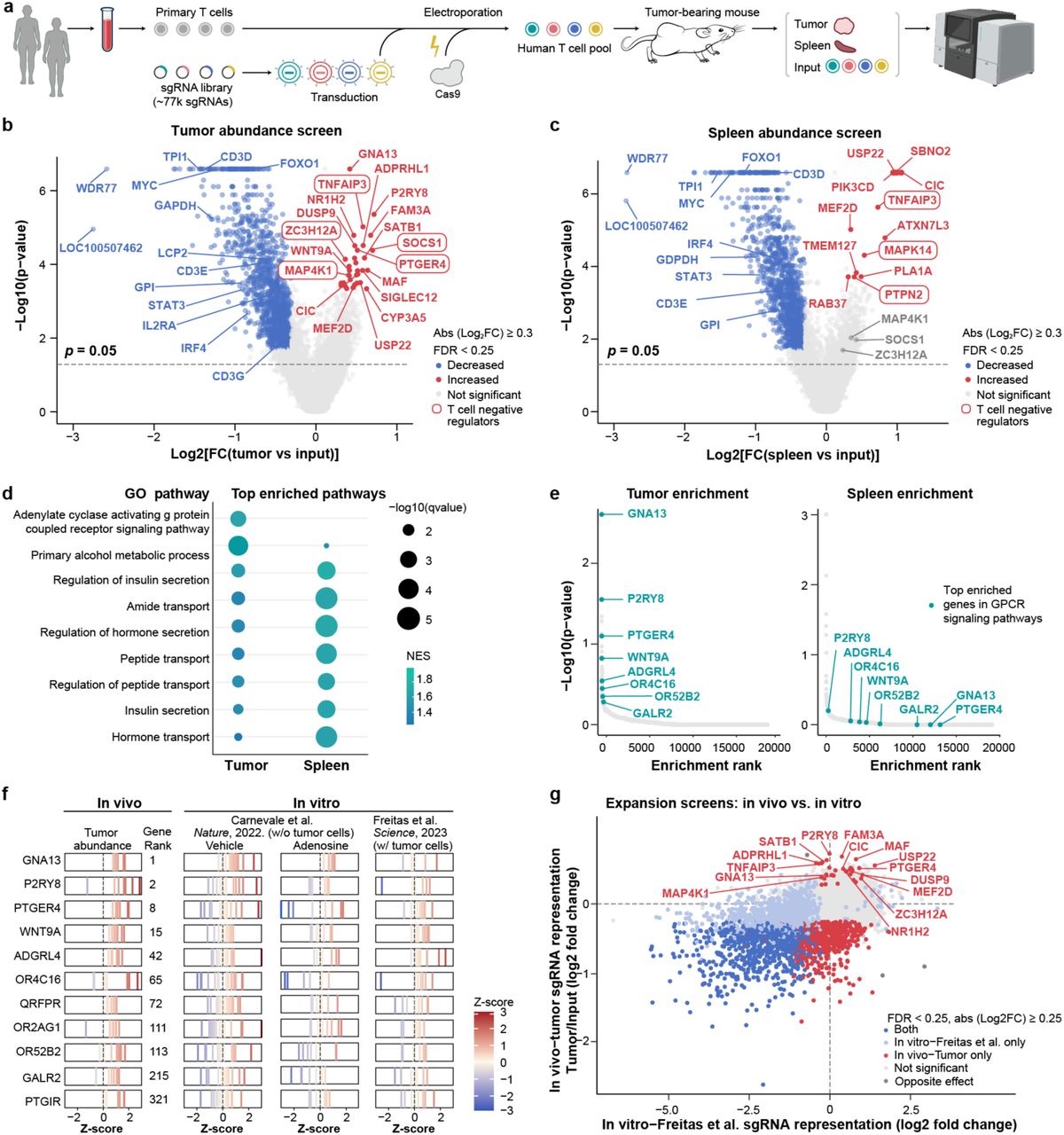
To uncover genetic factors that can improve T cell function within solid tumors, the researchers conducted two complementary genome-wide CRISPR knockout (KO) screens using human T cells.
Screen 1: Enhancing T Cell Infiltration into Tumors
In the first screen, a genome-scale CRISPR KO approach was applied to primary human T cells from two donors using the Brunello sgRNA library. After adoptive transfer into tumor-bearing mice, sgRNA abundance was tracked in tumor-infiltrating T cells (TILs) and splenic T cells to identify regulators of intratumoral T cell accumulation.
One of the top hits was the P2RY8–Gα13–ARHGEF1 pathway, which was found to suppress T cell infiltration. Mechanistically, P2RY8 was shown to be activated by GGG, a metabolite secreted by tumor cells, which inhibited T cell migration. Knocking out P2RY8 led to:
- Enhanced in vitro migration of T cells
- Increased in vivo tumor infiltration
- Modest but consistent improvements in tumor control
Interestingly, P2RY8 is human-specific and has no murine ortholog, highlighting the critical importance of using humanized models in solid tumor immunotherapy research.
Screen 2: Improving T Cell Effector Function Under Suppression
The second screen focused on effector function—specifically, IFN-γ production by TILs. T cells were isolated from tumors and sorted into high vs. low IFN-γ-producing subsets, and their CRISPR profiles compared to identify genes regulating cytokine output within the immunosuppressive tumor microenvironment (TME).
This screen identified GNAS, encoding the Gαs protein, as a major suppressor of T cell effector function. GNAS is a downstream node of several inhibitory G protein–coupled receptors (GPCRs), including:
- ADORA2A (adenosine A2A receptor)
- EP2 (prostaglandin E2 receptor)
- GPR65
- β-adrenergic receptors
GNAS KO T cells demonstrated remarkable resistance to multiple TME stressors:
- Maintained cytokine production under acidic pH, adenosine, prostaglandin E2, and β-adrenergic signals
- Preserved metabolism, viability, and IFN-γ output
- Achieved superior tumor control across multiple xenograft models, including A375 (melanoma), A549 (lung), MES-SA (sarcoma), and AsPC-1 (pancreatic)
Furthermore, GNAS-deficient T cells exhibited durable memory responses, suggesting long-term therapeutic benefit.
Combinatorial Editing: Dual Targeting of P2RY8 and GNAS
Building on the individual screens, the researchers next tested whether simultaneous disruption of trafficking and effector suppression pathways could further improve T cell performance. They engineered T cells with a dual knockout (KO) of P2RY8—to enhance migration—and GNAS—to confer resistance to immunosuppressive signaling within the tumor microenvironment.
The combined approach produced striking results:
- Greater T cell infiltration into tumors, reflecting improved trafficking
- Robust resistance to the suppressive effects of adenosine, prostaglandins, acidic pH, and β-adrenergic signaling
- Superior tumor clearance, achieving 4 of 6 complete responses in the stringent CD19⁺ A549 lung cancer model
These findings suggest that combinatorial editing of complementary pathways can unlock additive or even synergistic gains in T cell therapy for solid tumors, potentially overcoming two of the biggest barriers—poor trafficking and TME-induced dysfunction—at once.
Significance and Broader Impact
This study represents a landmark achievement—the first-ever genome-wide in vivo CRISPR screen performed directly in human T cells within solid tumors. By leveraging a physiologically relevant, tumor-bearing humanized model, the researchers identified targets that are often missed in traditional in vitro screens, highlighting the power of studying T cells in their true tumor context.
What makes this approach particularly impactful is its broad applicability. The identified targets and mechanisms can potentially improve performance across multiple T cell therapy platforms, including:
- CAR-T cells
- TCR-engineered T cells
- Tumor-infiltrating lymphocytes (TILs)
Beyond just identifying new therapeutic targets, the study also introduces a scalable and flexible platform for T cell engineering. This includes screening tools for:
- Loss-of-function (via CRISPR knockout)
- Gain-of-function (via ORF libraries)
- Multiplexed perturbations, such as base editing and combinatorial genetic engineering
In essence, this work lays the foundation for the next generation of engineered T cells—those with enhanced trafficking, persistence, and resilience against immunosuppression. It opens up new therapeutic avenues and provides a blueprint for how functional genomics can be harnessed to overcome the unique barriers of treating solid tumors.
Key Takeaway Messages
- P2RY8 limits T cell migration into tumors by sensing tumor-secreted GGG.
- GNAS (Gαs) integrates multiple suppressive GPCR signals in the TME; its deletion preserves T cell function.
- Dual gene editing of trafficking and resistance nodes may offer next-generation T cell therapy design.
- This platform is a major technological leap in modeling T cell biology in vivo, advancing cell therapy engineering.
 You Can Read the Full Article Here.
You Can Read the Full Article Here.