Immunotherapy, particularly immune checkpoint inhibition, has revolutionized modern oncology by achieving durable remissions across a wide range of tumor types. Despite these transformative outcomes, only 20–30% of patientsexperience sustained benefit, underscoring a critical challenge in accurately predicting who will respond to treatment. The clinical and economic implications are substantial—given that the global ICI market is projected to surpass $189 billion by 2032, the urgency for precise, clinically actionable predictive tools has never been greater.
In this comprehensive and forward-looking review, Oisakede et al. explore the rapidly advancing field of predictive modeling for immunotherapy response, encompassing biomarker-driven strategies, artificial intelligence and machine learning algorithms, mechanistic modeling, and multi-modal integration frameworks. Drawing from more than 200 studies, the authors deliver one of the most extensive comparative analyses to date, evaluating predictive accuracy, clinical applicability, and translational readiness across emerging methodologies.
Key Findings and Conceptual Advances
Limitations of single biomarkers
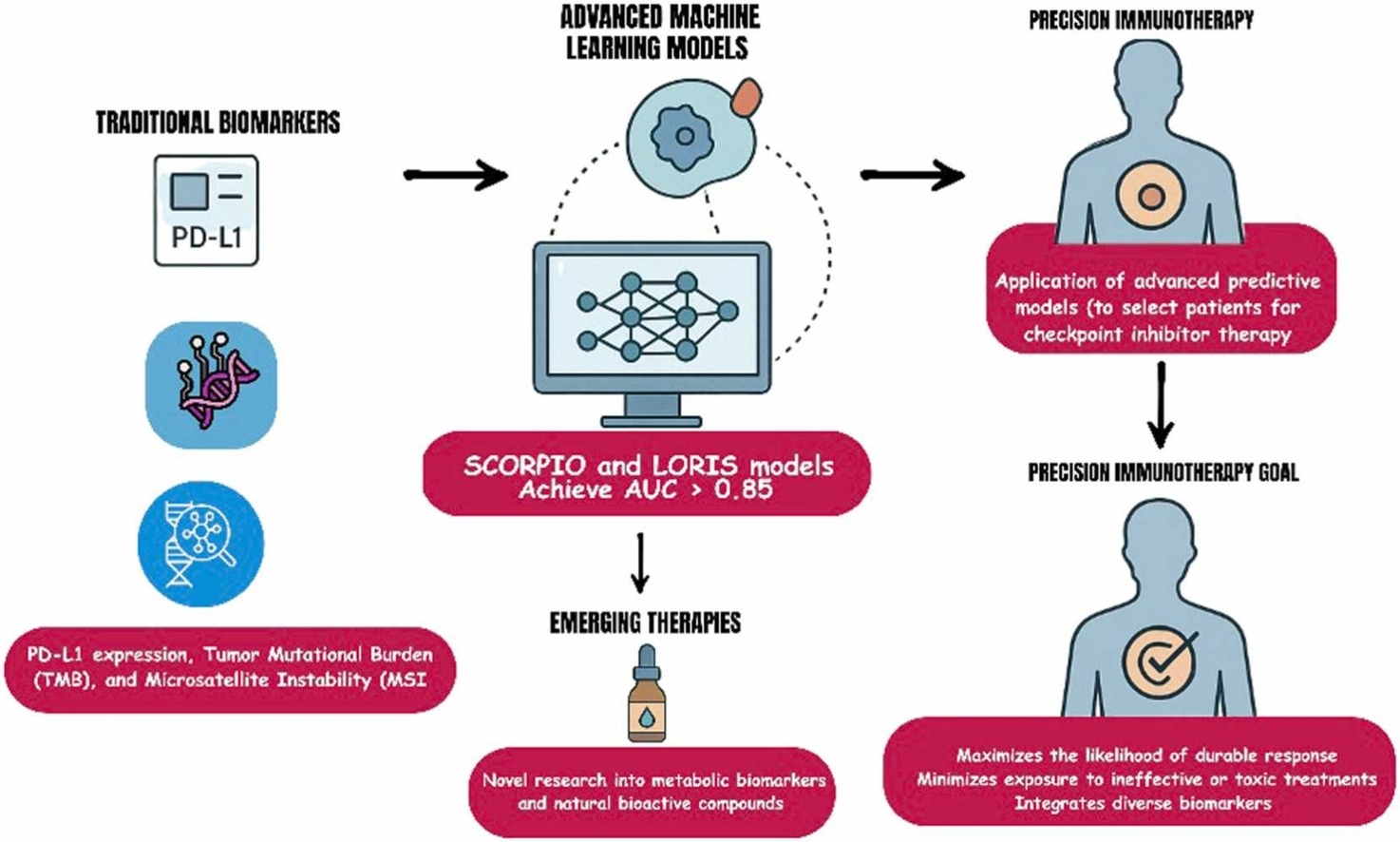
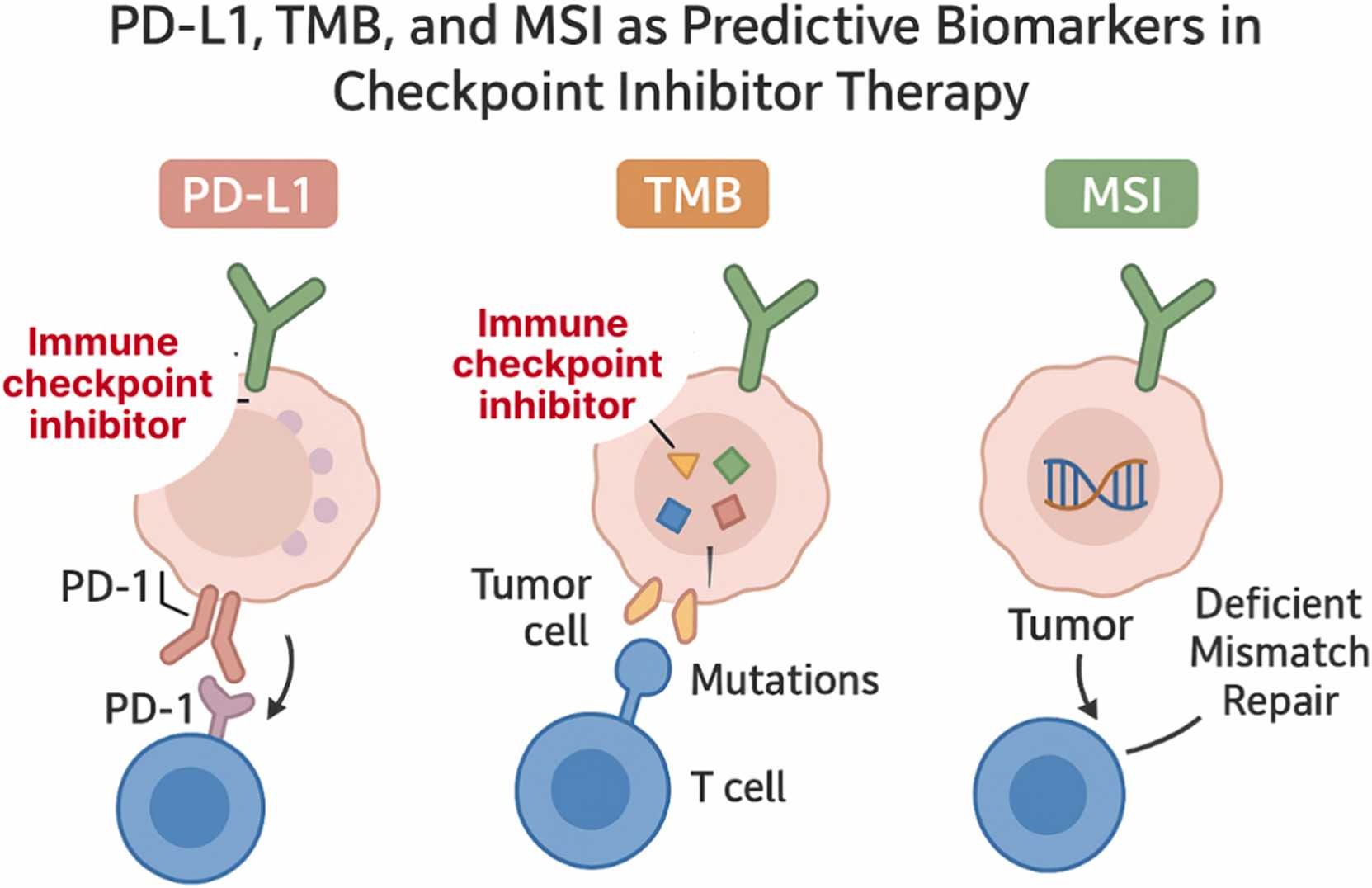
Traditional biomarkers—such as PD-L1 expression, tumor mutational burden (TMB), and microsatellite instability (MSI)—have long served as the foundation for selecting patients who might benefit from immune checkpoint inhibitors. However, their predictive accuracy remains limited.
PD-L1 expression, for instance, is predictive in only about 29% of FDA-approved indications, and both TMB and MSI show highly variable reliability across cancer types. These inconsistencies stem from biological heterogeneity, differences in testing platforms, and the complex interplay between tumor and immune factors.
The authors highlight that predicting immunotherapy response cannot rely on any single molecular marker. Instead, multi-parametric models—those integrating molecular, immunologic, and spatial data—are needed to capture the full biological context of the tumor microenvironment (TME).
The rise of metabolic biomarkers
Beyond genomics, metabolic reprogramming has emerged as a critical determinant of immune evasion and treatment resistance.
Tumors with elevated expression of glucose transporters GLUT1 and GLUT3 exhibit enhanced glycolysis, creating an acidic microenvironment that suppresses T-cell activity. This glucose competition between tumor and immune cells not only limits immune effector function but also promotes immune exhaustion.
Incorporating these metabolic biomarkers into predictive models could refine patient stratification—particularly for tumors that are both metabolically active and immunologically “cold.” Such integrated models could help identify patients who may benefit from therapies that target both metabolism and immune regulation.
Artificial intelligence outperforms traditional biomarkers
Artificial intelligence (AI) and machine learning (ML) now represent the fastest-growing frontiers in predictive oncology. These approaches are capable of integrating high-dimensional clinical, molecular, and imaging data to uncover complex patterns not visible to human observers.
- The SCORPIO model, developed at Memorial Sloan Kettering Cancer Center, analyzed data from nearly 10,000 patients across 21 cancer types and achieved an AUC of 0.76 for predicting overall survival—significantly outperforming PD-L1 and TMB.
- The LORIS model, based on six routine clinical and genomic parameters (age, albumin, neutrophil-to-lymphocyte ratio, TMB, prior therapy, and cancer type), achieved 81% predictive accuracy and showed strong external validation across multiple international cohorts.
- Deep learning approaches applied to histopathology images have further advanced predictive precision, enabling automated assessment of PD-L1 expression and tumor-infiltrating lymphocytes (TILs) with AUC values exceeding 0.9 in controlled research settings.
Despite these advances, one major challenge persists: external validation. Many AI models perform exceptionally well within the institution where they were developed but fail to maintain accuracy when tested on independent patient populations—a problem the authors refer to as the “validation gap.”
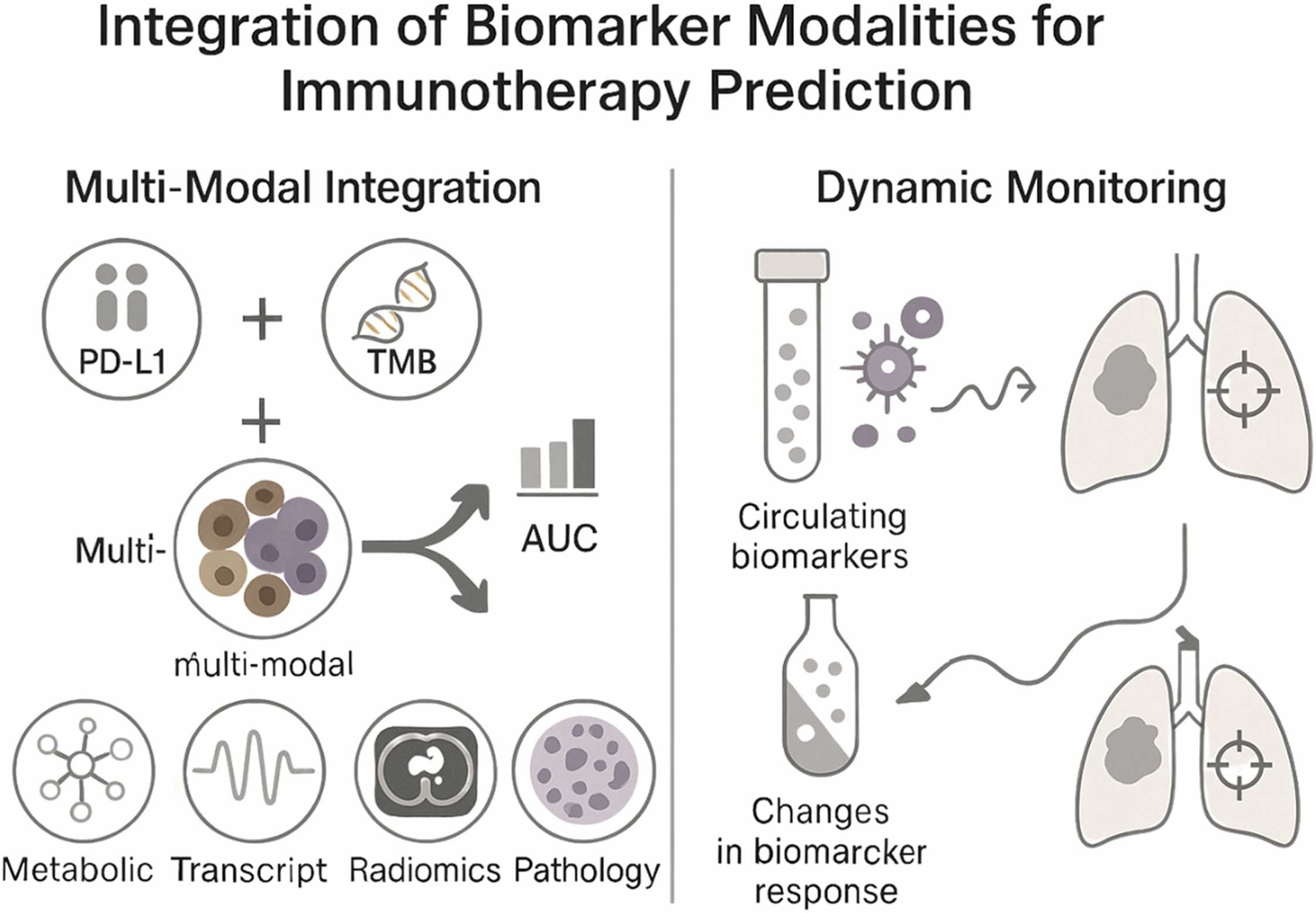
Integrating multi-modal data for precision prediction
Combining multiple types of data—genomic, spatial, clinical, and metabolic—has proven far more effective than using single-modality biomarkers.
These multi-modal frameworks have achieved AUC values above 0.85 in several cancers, outperforming traditional metrics. For example, integrating PD-L1 expression, TMB, and immune cell infiltration patterns improved predictive power in non–small cell lung cancer and melanoma.
Modern spatial profiling technologies, such as multiplex immunofluorescence and digital spatial transcriptomics, now reveal how immune and tumor cells are organized within the TME. This spatial information often correlates more strongly with treatment response than bulk biomarker measurements, underscoring the importance of tumor architecture in predicting therapeutic outcomes.
Dynamic and mechanistic modeling approaches
A new generation of mathematical and systems biology models aims to simulate tumor–immune interactions in real time. These models capture key parameters—such as tumor growth kinetics, immune infiltration, and checkpoint blockade dynamics—to forecast treatment outcomes and understand resistance mechanisms.
Early studies show promising results: some of these mechanistic models can classify responders versus non-responders with up to 81% accuracy in pilot validation cohorts. Although still in early stages, these models complement data-driven AI approaches by offering a mechanistic understanding of immune dynamics, which could ultimately guide personalized dosing, combination strategies, and treatment adaptation.
Natural Compounds and Metabolic–Immune Crosstalk
A novel section of the review explores the integration of natural bioactive compounds—such as thymoquinone (Nigella sativa), cucurbitacins (Cucurbitaceae), and organosulfur compounds (garlic derivatives)—which have demonstrated immunomodulatory and metabolic reprogramming effects in preclinical studies. These agents may augment ICI efficacy by:
- Reducing tumor glycolysis and acidosis;
- Enhancing T-cell function and cytokine production;
- Modulating epigenetic regulators such as HDAC2;
- Inhibiting key pro-survival pathways (NF-κB, JAK2/STAT3, PI3K/AKT).
Such multi-target actions highlight a potential role for metabolic–immune combinatorial therapy, though clinical validation remains limited.
Implementation Challenges
The review identifies three persistent barriers that hinder translation from research to practice:
- Validation and reproducibility: promising models rarely replicate performance outside their development cohort.
- Data standardisation: inconsistencies in biomarker assays, imaging platforms, and sequencing pipelines undermine generalisability.
- Healthcare integration: lack of interoperability between predictive algorithms and clinical information systems delays implementation in real-world oncology workflows.
The authors call for international standardisation frameworks, similar to those of the Global Alliance for Genomics and Health (GA4GH), to harmonise data collection, model validation, and AI governance in oncology.
Significance
This landmark review represents one of the most comprehensive syntheses of predictive model development in immuno-oncology. It bridges traditional pathology and next-generation computational science, highlighting the need for multidisciplinary collaboration among pathologists, oncologists, data scientists, and regulatory bodies.
By systematically evaluating every major class of predictive approach—from PD-L1 scoring to AI integration—this work outlines a roadmap for clinically implementable, validated, and interpretable models capable of guiding patient selection and optimizing immune checkpoint inhibitor therapy.
Key Takeaway Messages
- ICIs benefit only a minority of patients; precision prediction is critical for therapeutic success.
- AI and multi-modal models outperform traditional biomarkers, but external validation remains the main translational bottleneck.
- Integration of metabolic and spatial biomarkers provides new biological dimensions for prediction.
- Natural bioactive compounds may enhance checkpoint inhibitor efficacy via metabolic and immune modulation.
- Future success depends on global standardisation, real-time adaptive modeling, and clinically interpretable AI integration.
You Can Read Full Article Here


