Lewis Cantley, Professor of Cell Biology at Harvard Medical School, shared a thread on X: .
“We are delighted to share the Tyrosine Kinase Library, now in Nature. Tyrosine predictions and enrichment analysis are now available.
Work led by Jared Johnson and Tomer M. Yaron-Barir, together with The Yaffe Lab and Turk Lab!
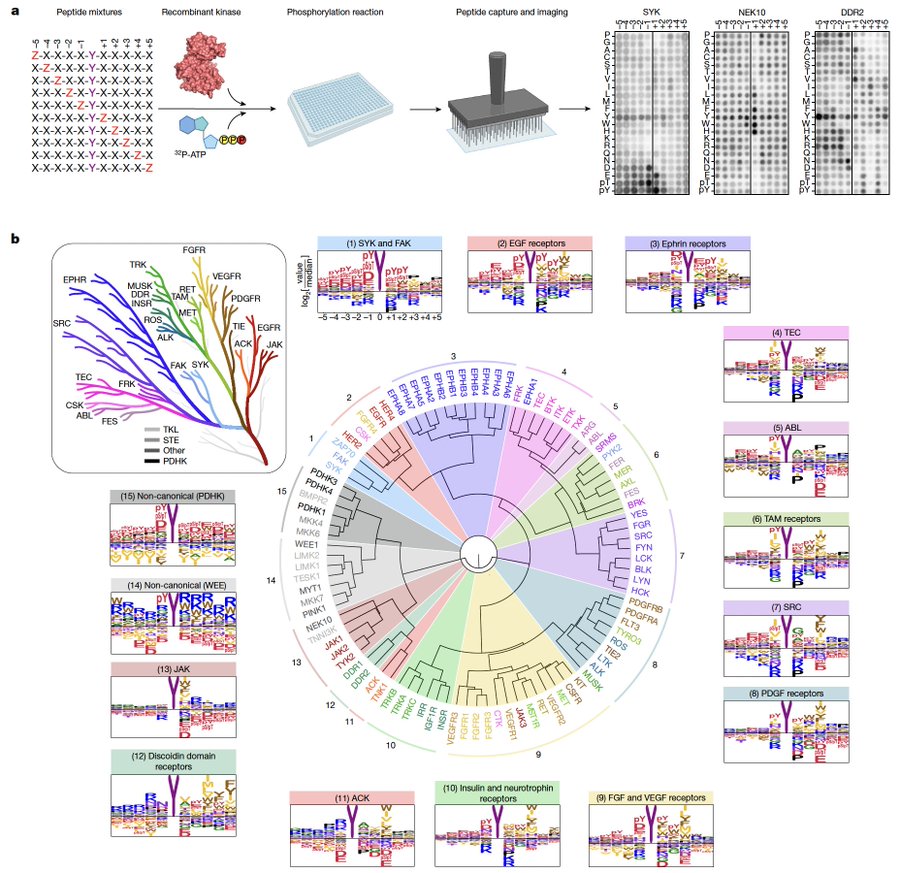
Using synthetic peptide libraries, we characterized the biochemical substrate specificity of every human tyrosine kinase – 78 canonical and 15 non-canonical kinases, creating the tyrosine kinome wheel:
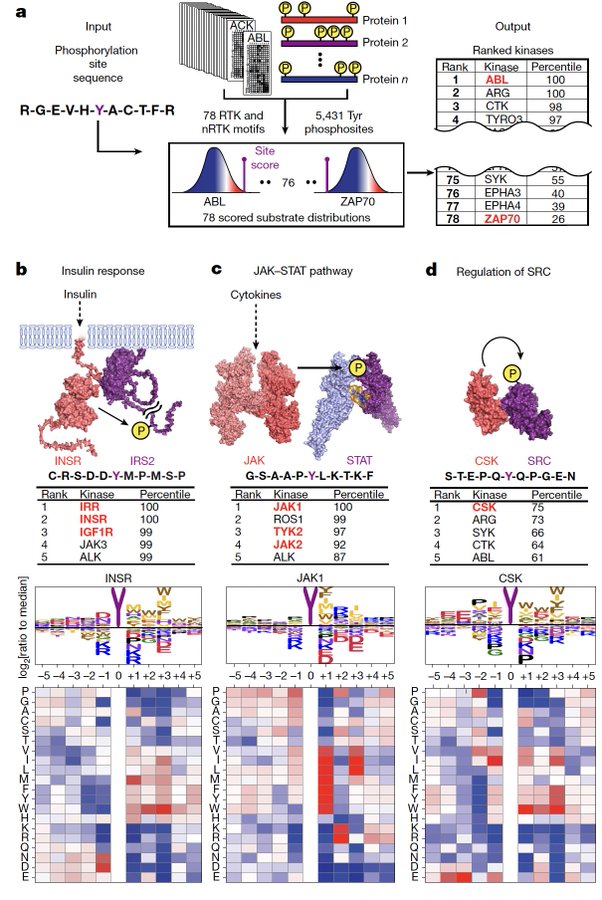
Utilizing our scoring algorithm, we can identify the upstream kinases for well-studied kinase-substrate relationships in the insulin, cytokine, and SRC signaling pathways:
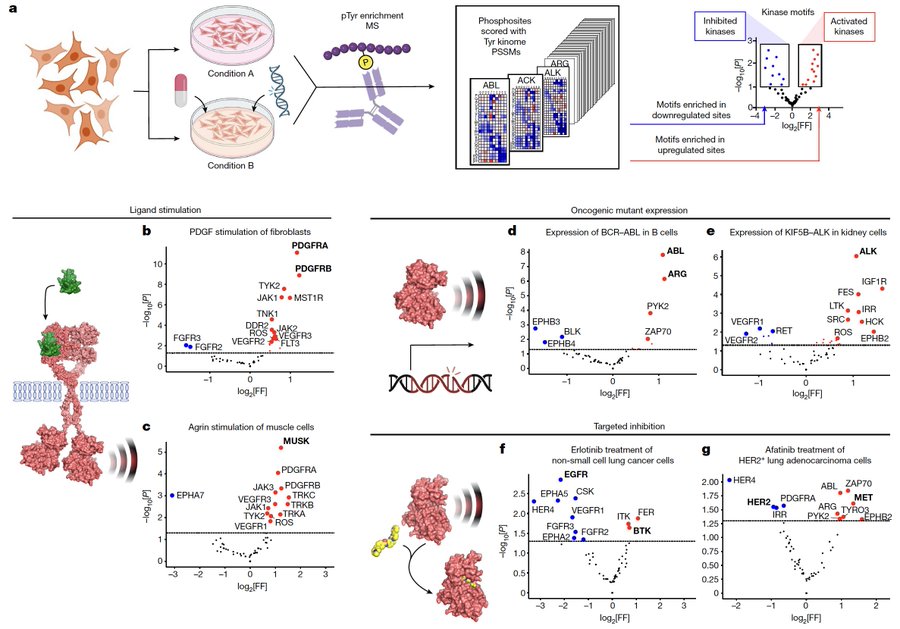
Based solely on the sequence context of the detected phosphosites, our enrichment algorithm identifies the regulated kinases in high-throughput tyrosine phosphoproteomics experiments:
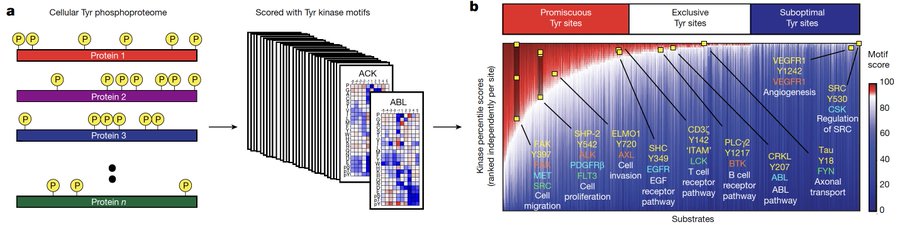
We also annotated the human tyrosine phosphoproteome and reclassify phosphorylation sites into (1) promiscuous (liked by many kinases), (2) exclusive (liked by a few), and (3) suboptimal (poorly matching the motifs of every conventional tyrosine kinase):
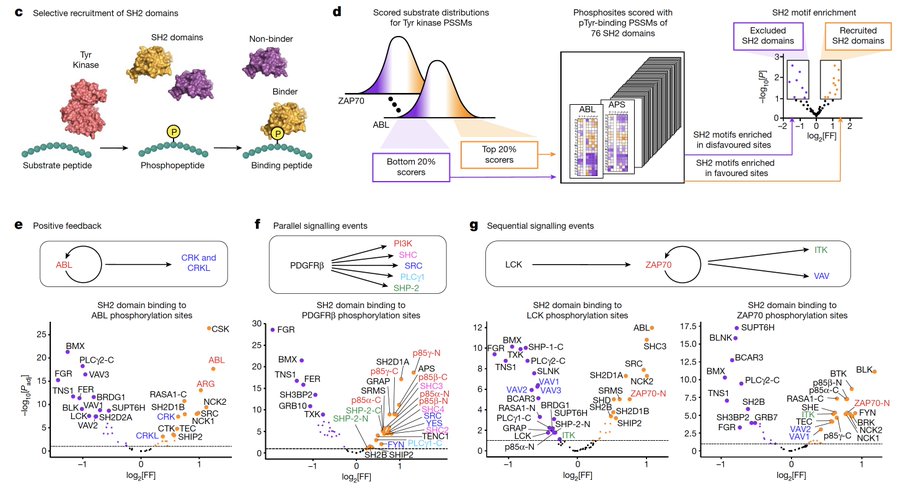
We then integrated our motif library with SH2 domain sequence specificities, and fascinatingly the topology of known tyrosine signaling networks emerges:
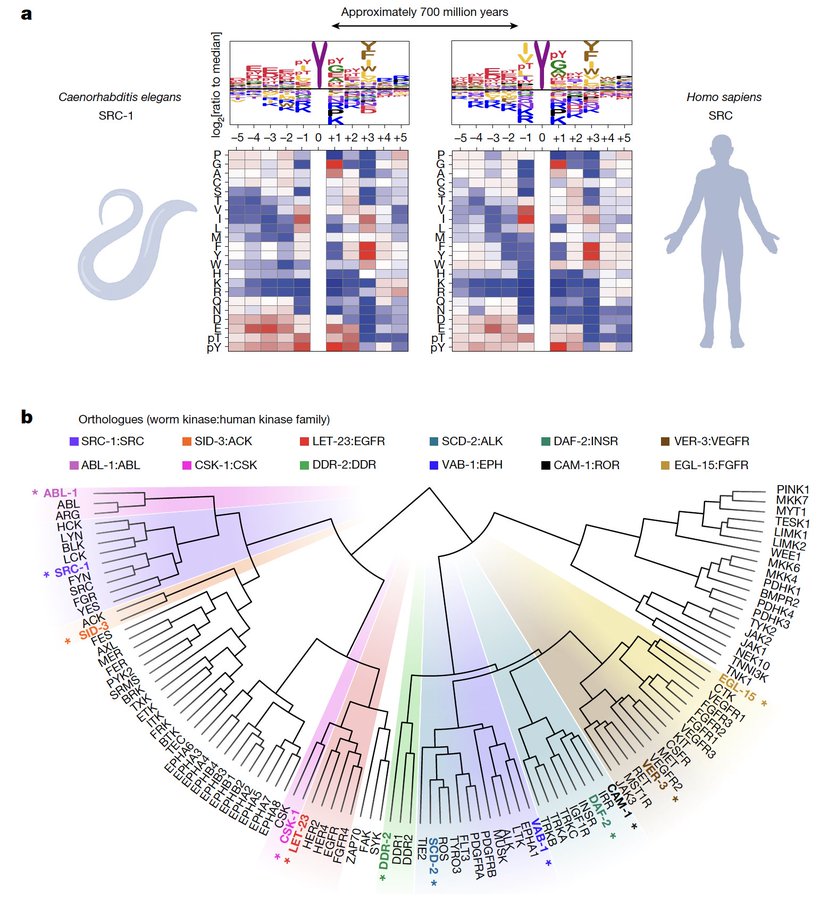
Not a lot has changed over the past 700 million years! Finally, we characterized multiple C. elegans orthologues and found that the motif specificity of worm tyrosine kinases is surprisingly similar to that of humans:
Coming very soon: The Kinase Library Python package, as well as new features for substrate predictions and enrichment analysis, including continuous enrichment methods, phosphopriming, lysine acetylation and more!”
Source: Lewis Cantley/X