BRAF Isoforms and Resistance to MAPK Inhibitors in Melanoma
Authors: Francisco Aya, Pablo Lanuza-Gracia, Abel González-Pérez, Sophie Bonnal, Estefania Mancini, Nuria López-Bigas, Ana Arance, Juan Valcárcel
Published in Cell Reports on April 23, 2024 .
Introduction:
Resistance to MAPK inhibitors (MAPKi), such as BRAF and MEK inhibitors, is a major challenge in treating BRAF-mutant melanoma. The production of alternative BRAF mRNA isoforms (altBRAFs) has been associated with up to 30% of cases of resistance to BRAF inhibitor monotherapy.
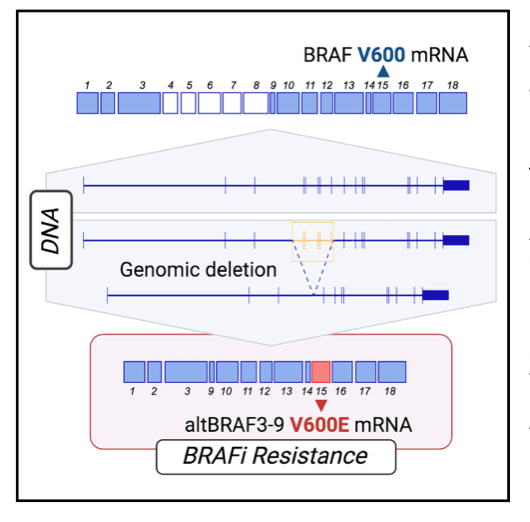
Previously, altBRAFs were thought to be generated by alternative splicing, leading to proposals for splicing modulation as a therapeutic strategy. This study challenges this view and provides evidence that altBRAFs are primarily caused by genomic deletions in the BRAF gene.
Design/Methods:
The researchers utilized different transcriptome datasets, including RNA-seq data from melanoma cell lines and patient tumors, to identify and quantify the expression of altBRAFs.
They employed whole-genome sequencing (WGS) and PCR amplification followed by Sanger sequencing to detect genomic deletions in the BRAF locus. In vitro models of altBRAF-mediated melanoma resistance were used to study the mechanisms of altBRAF generation.
Key Findings:
- altBRAFs were detected not only in BRAF-mutant and MAPKi-resistant samples but also in treatment-naive and BRAF wild-type melanoma samples, suggesting that altBRAFs are not solely associated with MAPKi resistance.
- In resistant melanoma cell lines, altBRAFs were exclusively expressed from the BRAF V600E allele, indicating a cis-acting genetic alteration rather than a trans-acting splicing factor alteration.
- Whole-genome sequencing and PCR amplification revealed cell line-specific genomic deletions in the BRAF locus, precisely encompassing the skipped exons responsible for generating different altBRAF isoforms.
- Evidence of genomic rearrangements was also found in patient tumor samples, which was consistent with the observed altBRAF expression profiles.
Key Highlights:
- altBRAFs are generated through genomic deletions, not alternative splicing.
- This challenges the prevailing paradigm of altBRAFs being produced by competition between alternative splice sites.
- Genomic deletions were detected in resistant cell lines and patient tumors, explaining the generation of specific altBRAF isoforms.
- altBRAFs were found in treatment-naive and BRAF wild-type tumors, suggesting a broader relevance beyond MAPKi resistance.
Key Takeaway Messages:
- The generation of altBRAFs in melanoma is primarily driven by genomic deletions in the BRAF locus rather than alternative splicing events.
- This finding has significant implications for understanding the resistance mechanisms to MAPKi and challenges the proposed therapeutic strategy of using splicing modulators to overcome resistance.
- The detection of altBRAFs in treatment-naive and wild-type BRAF tumors expands the potential clinical relevance of these BRAF isoforms beyond MAPKi resistance.
- Further investigation is warranted to explore the effects of type II RAF inhibitors in tumours harbouring dimer-enhancing genomic deletions in BRAF, regardless of BRAF mutational status.
Summary by Amalya Sargsyan, MD


