Livnat Jerby, Assistant Professor at Stanford University, posted on X:
“Our new paper is out in Nature Immunology. Applying spatial transcriptomics at subcellular resolution to 130 ovarian cancer tumors, followed by data-driven experimental design and high content CRISPR screens to map immune evasion in ovarian cancer
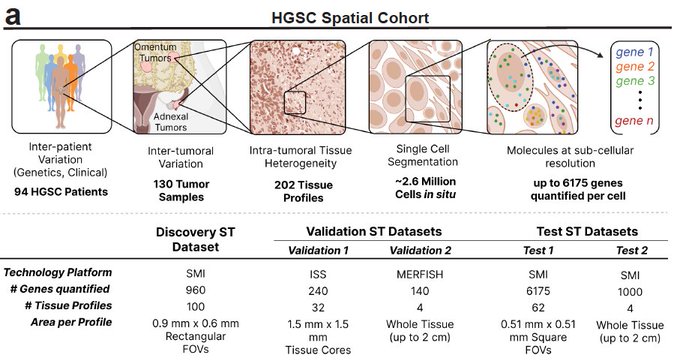
Immune responses depend, shape and are shaped by spatial organization. To study tumor immunology across large patient populations, we applied spatial transcriptomics to >2.5 million cells from 130 ovarian cancer tumors from 94 patients, with hundreds to thousands of genes measured in situ at single molecule resolution.
To figure out cause and effect we: (1) identified cell state regulators by integrating the spatial data with the patients’ tumor genetics. (2) Performed high content CRISPR screens, allowing us to perform dozens of experiments at a time with data-driven experimental design.
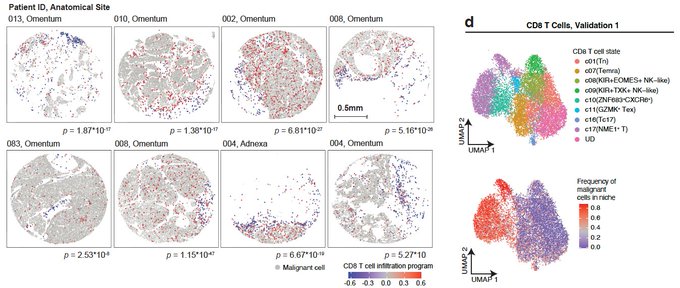
One of the biggest questions that we wanted to address was: What is holding T cells from getting recruited and infiltrating into the tumor? We found that T cells that are trapped/sequestered to the stroma (white areas) are in a naïve/memory state (blue), while effector and cytotoxic T cells (red) are next to malignant cells (grey).
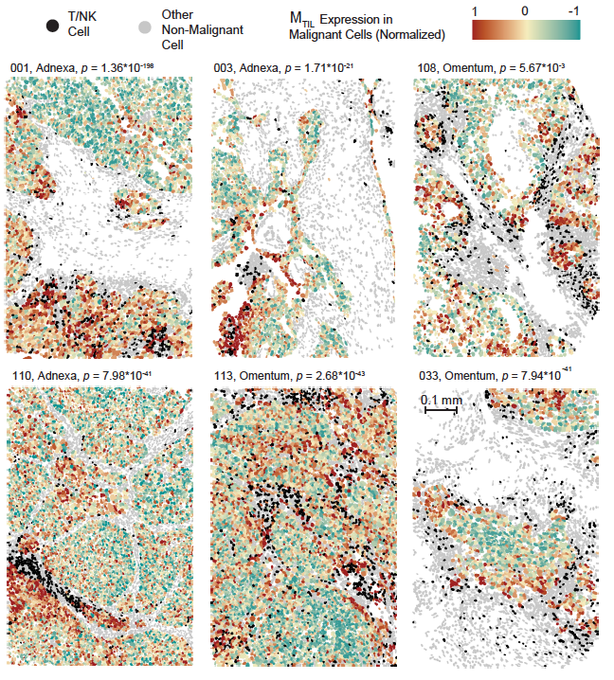
So, effector T cells and Natural Killer (NK) cells (black) are getting into the tumor, BUT they are spatially confined and preferentially co-localize only with a subset of malignant cells. What is unique about this malignant cell subset? Their transcriptional state.
This malignant cell state includes dozens of co-regulated genes, including up-regulation of chemokines (e.g., CXCL16), and cell adhesion molecules, down-regulation of Wnt signaling, epigenetic regulators (DNMT3A, HDAC1/11/4/5), insulin signaling genes and many more.
We hypothesized that this is an immunogenic cell state. Namely, a transcriptional state that “allows” malignant to be recognized and attacked by T cells / NK cells
Is the immunogenic cell state related to intrinsic properties of malignant cells? Yes. It is strongly associated with copy number alterations both in cis and in trans and varies across patients, even when considering only niches without any T/NK cells.
Using this cell state’s gene deletions/amplifications we can predict whether a tumor is immunologically ‘hot’ or ‘cold’ (AUROC>0.8), suggesting that the genetic basis of immunogenicity goes beyond neoantigens.
These principles of tumor organization and cell-to-environment interactions are consistent and generalizable (even across cancer types), and the immunogenic malignant cell state predicts whether a patient will respond to immunotherapy.
So, can we activate the immunogenic cell state? Yes. We performed Perturb-Seq CRISPR screens in ovarian cancer cells and identified gene knockouts that activate/repress it.
How did we decide which genes to target in our Perturb-seq screen? By using previous Perturb-Seq screens! This was critical because many of the regulators/hits were not a part of the immunogenic cell state itself.
In our screen we identified KOs that activate the immunogenic state and (as predicted) sensitize the cancer cells to T cell / NK killing. Top hits are the epigenetic regulator ACTR8 and insulin sensing repressor PTPN1, opening new avenues to unleash anti-tumor immune responses.
This study was led by three talented members of my lab Christine Yiwen Yeh, Karmen Aguirre, and Olivia Laveroni, and in collaboration with Brooke E. Howitt. A big thank you to all lab members and co-authors, Mike Bassik, Sylvia Katina Plevritis, and Michael Snyder.
There is a lot more in the paper, so hope you go and check it out!”
Source: Livnat Jerby/X