
Sanam Loghavi: Data-Driven Insights Refine Myeloid Neoplasm Classification
Sanam Loghavi, Associate Professor of Pathology at MD Anderson Cancer Center, Medical Director of ECOG-ACRIN Leukemia Bank and Editor in Chief of Image Bank at the American Society of Hematology, shared a post on X:
“New paper!
Myeloid neoplasms with a data-driven twist!
Our latest in addresses classification gaps in the world of myeloid neoplasms, led by Curtis Lachowiez in collaboration with Elli Papaemmanuil, Elsa Bernard and a huge team of amazing friends and collaborators.
We tackled 3 major disease categories։
- SF3B1 mut MDS
- NPM1 mut AML
- CMML (the oligomonocytic type)
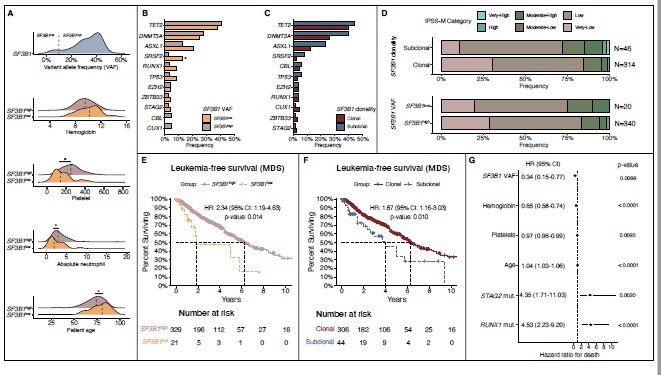
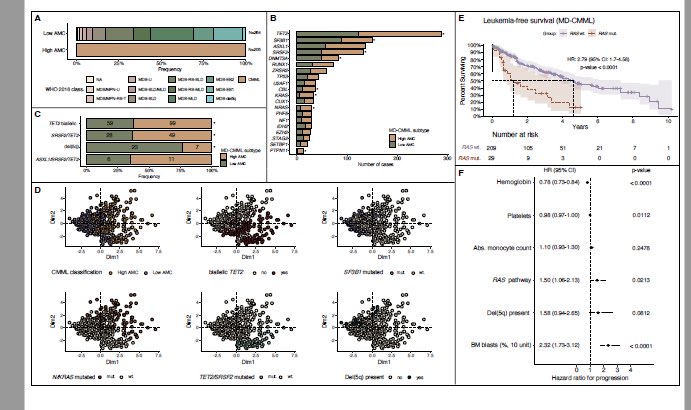
SF3B1-m MDS: Clonal burden (size) matters!
SF3B1 VAF ≥10% vs <10% changes the game—co-mutations, clonality, and survival!
Low VAF/subclonal SF3B1:not so great LFS , co-mutations, IPSS-M risk
Clonality is key!
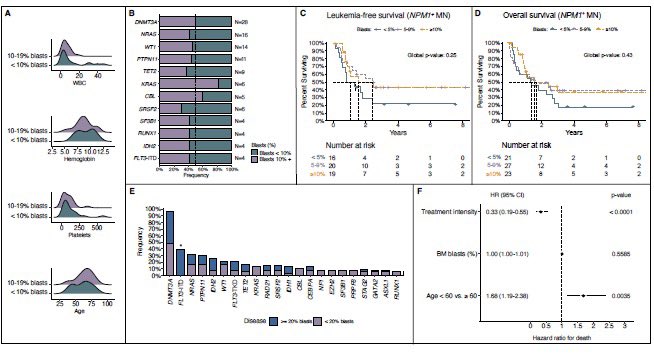
NPM1-mutated AML: Blast count? Irrelevant.
NPM1 mutations define AML regardless of blasts (5% WHO and 10% ICC cutoffs both irrelevant)
Same survival, same genetics—time to treat these as one entity.
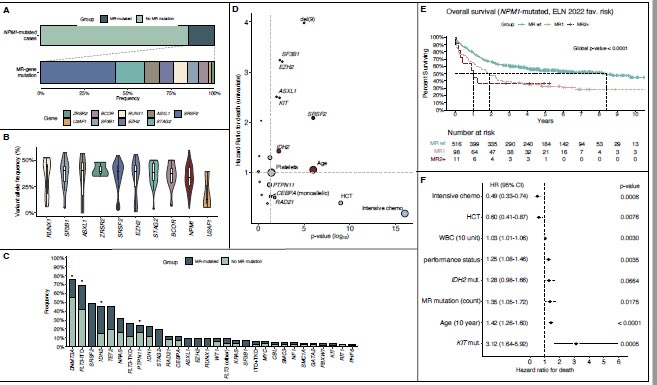
NPM1 + MR mutations: (SRSF2 tops the list), currently favorable by ELN 2022 In our analysis MR mutations+NPM1= worse survival than NPM1 without MR mutations. Esp if >1 MR mut.
O-CMML (oligomonocytic) classification needs some TLC!
We need to incorporate CMML type gene signatures for O-CMML diagnosis… otherwise you’ll see a bunch of MDS misclassified as CMML.
More genetic signatures, please!
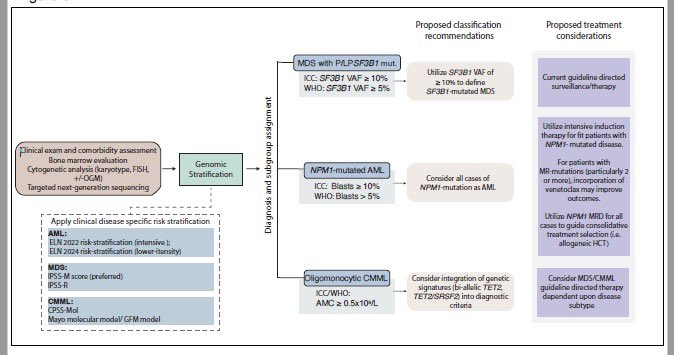
Data-driven classifications FTW! Our recommended workflow for molecularly precise diagnosis and treatment in MDS, AML, and CMML.
Let’s rethink those thresholds!
A tremendous to our awesome team of collaborators who helped by along the way and contributed pt cases to help us demonstrate these points.
Guillermo Montalban-Bravo, Jad Othman, Richard Dillon, Ak Eisfeld, and the entire IWG and UKNCRI colleagues.
To our reviewers (you know who you are) for improving the paper.
And to Tanya Bondar and the entire Blood Cancer Discovery for their help and guidance throughout the review and publication process.”
Title: Multimodal and Data-Driven Assessment of Myeloid Neoplasms Refines Classification across Disease States
Journal: Blood Cancer Discovery
Authors: Curtis A. Lachowiez, Georgios Asimomitis, Elsa Bernard, Sean M. Devlin, Yanis Tazi, Maria Creignou, Ulrich Germing, Norbert Gattermann, Amanda Gilkes, Ian Thomas, Lars Bullinger, Konstanze Döhner, Luca Malcovati, Jad Othman, Richard Dillon, Ann-Kathrin Eisfeld, Deedra Nicolet, Ghayas C. Issa, Naval Daver, Tapan M. Kadia, Courtney D. DiNardo, Farhad Ravandi, Guillermo Garcia-Manero, Guillermo Montalban-Bravo, Nigel Russell, Mario Cazzola, Hartmut Döhner, Brian J.P. Huntly, Robert P. Hasserjian, Eva Hellström-Lindberg, Elli Papaemmanuil, Sanam Loghavi

Later she added to this post:
“You can use this QR code or link to access our recent paper on classification of myeloid neoplasms in Blood Cancer Discovery for free for the duration of Society of Hematologic Oncology next week.”

More posts featuring Sanam Loghavi on OncoDaily.
-
Challenging the Status Quo in Colorectal Cancer 2024
December 6-8, 2024
-
ESMO 2024 Congress
September 13-17, 2024
-
ASCO Annual Meeting
May 30 - June 4, 2024
-
Yvonne Award 2024
May 31, 2024
-
OncoThon 2024, Online
Feb. 15, 2024
-
Global Summit on War & Cancer 2023, Online
Dec. 14-16, 2023
