The BARCODE1 study investigates the clinical utility of polygenic risk score (PRS)–based stratification in population-level prostate cancer screening. In light of the ongoing debate over PSA-based screening—due to concerns about false positives, overdiagnosis, and overtreatment—this prospective study aimed to evaluate whether targeting men at the highest genetic risk could improve detection of clinically significant prostate cancer while minimizing unnecessary interventions. By combining genomic profiling with standard diagnostic tools such as PSA testing and MRI, the study introduces a personalized screening model that may refine current approaches to early detection.
Title: Assessment of a Polygenic Risk Score in Screening for Prostate Cancer
Authors:
Jana K. McHugh, , Elizabeth K. Bancroft, Ph.D., Edward Saunders, M.Sc., Mark N. Brook, Ph.D., Eva McGrowder, Ph.D., Sarah Wakerell, M.Sc., Denzil James, M.Sc., Reshma Rageevakumar, M.Sc., Barbara Benton, R.N., Natalie Taylor, R.N., Kathryn Myhill, R.N., Matthew Hogben, R.N., Netty Kinsella, Ph.D., Aslam A. Sohaib, F.R.C.R., Declan Cahill, Stephen Hazell, F.R.C.Path., Samuel J. Withey, M.B., B.S., Naami Mcaddy, M.B., B.S., Elizabeth C. Page, M.Sc., Andrea Osborne, Sarah Benafif, Ph.D., Ann-Britt Jones, M.R.C.P., Dhruv Patel,
Dean Y. Huang, M.D., Kaljit Kaur, R.N., Bradley Russell, R.N., Ray Nicholson, R.N., Fionnuala Croft, R.N., Justyna Sobczak, R.N., Claire McNally, R.N., Fiona Mutch, R.N., Samantha Bennett, Lenita Kingston, Questa Karlsson, Ph.D., Tokhir Dadaev, M.Sc., Sibel Saya, Ph.D., Susan Merson, B.Sc., Angela Wood, Nening Dennis, B.Sc., Nafisa Hussain, B.Sc., Alison Thwaites, Syed Hussain, B.Sc., Imran Rafi, Ph.D., Michelle Ferris, M.D., Pardeep Kumar, Nicholas D. James, Ph.D., Nora Pashayan, Ph.D., Zsofia Kote-Jarai, Ph.D., and Rosalind A. Eeles, Ph.D.
Published in NEJM, April 2025
Background
Prostate cancer is the second most common cancer and a leading cause of cancer-related death among individuals assigned male at birth. In 2020 alone, it caused approximately 375,000 deaths globally. Despite this burden, there is no internationally standardized population-based screening program. While PSA testing is widely used, it has limitations due to false positives, overdiagnosis, and overtreatment. Long-term data, such as from the ERSPC trial, indicate a mortality reduction of up to 30% with PSA-based screening. Nonetheless, more precise risk stratification tools are urgently needed.
Prostat cancer is highly heritable, with 58% heritability in twin studies. While rare variants in genes like BRCA1/2 contribute to risk, a larger portion is attributable to common low-risk variants or SNPs. The cumulative effect of these SNPs can be quantified using a polygenic risk score (PRS). The BARCODE1 study aimed to prospectively evaluate PRS-based stratification in a population-level prostate cancer screening approach.
Methods and Study Design
BARCODE1 is a single-group, prospective study conducted across 69 primary care centers in the UK. Eligible participants were men aged 55–69 years, of European ancestry, without a history of prostate cancer. Participants mailed saliva samples for genotyping of 130 validated SNPs.
Those with a PRS in the ≥90th percentile, based on reference population data, were referred for further screening: PSA testing, multiparametric MRI, and transperineal prostate biopsy. MRI was interpreted using PI-RADS v2.1, and all biopsies were performed under local anesthesia. Cancers were classified per Gleason score and NCCN 2024 guidelines.
Sample Size
The target sample size was 5,000 to yield at least 500 men in the top 10% of PRS. It was estimated that this group would account for ~29% of prostate cancer cases.
Statistical Analysis
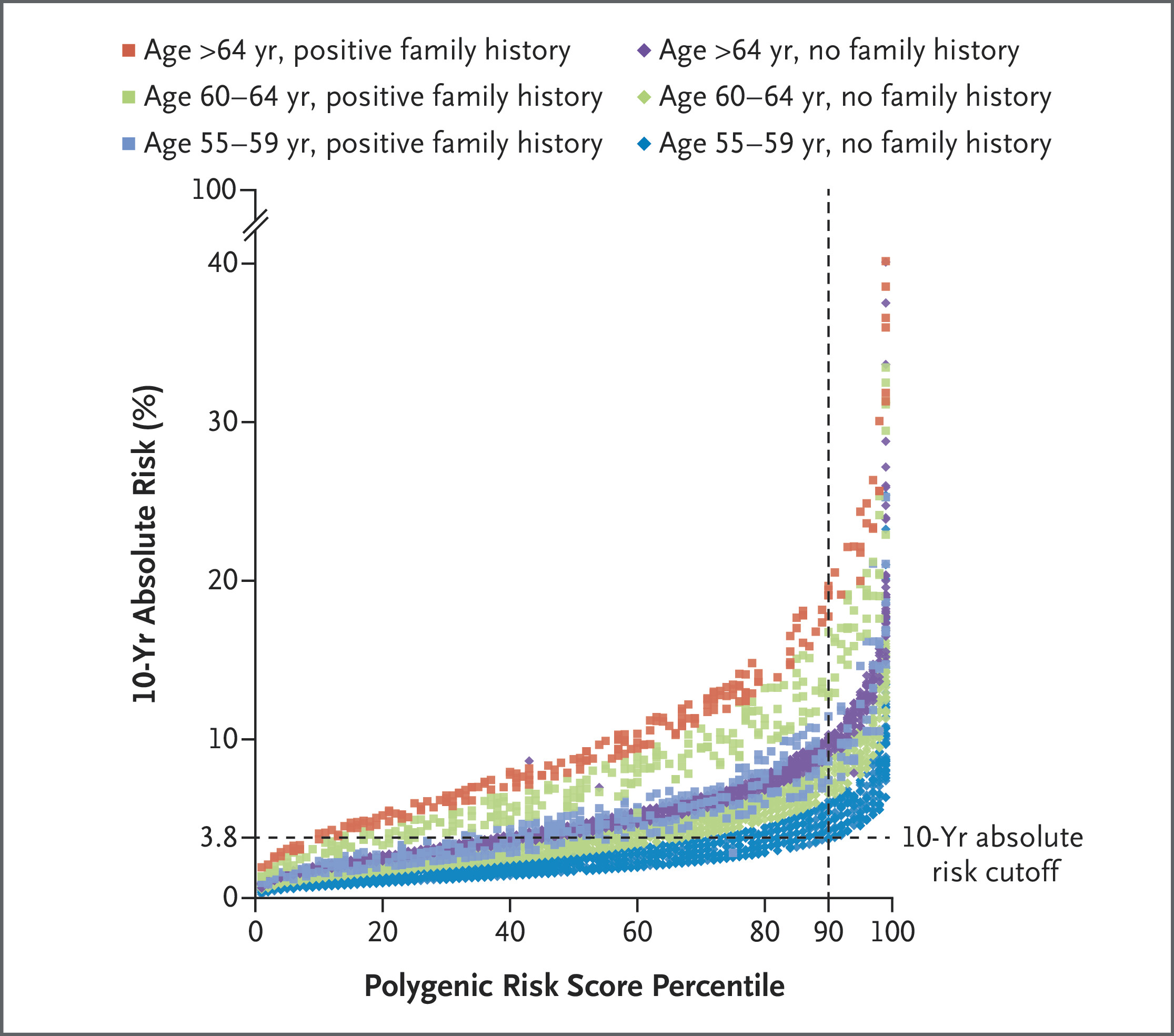
Outcomes included prostate cancer detection, positive predictive values (PPVs), Gleason scores, and overdiagnosis estimates. Logistic regression evaluated predictors of cancer detection. Ten-year absolute risk was calculated using the iCARE R package, integrating age, family history, and PRS.
Results
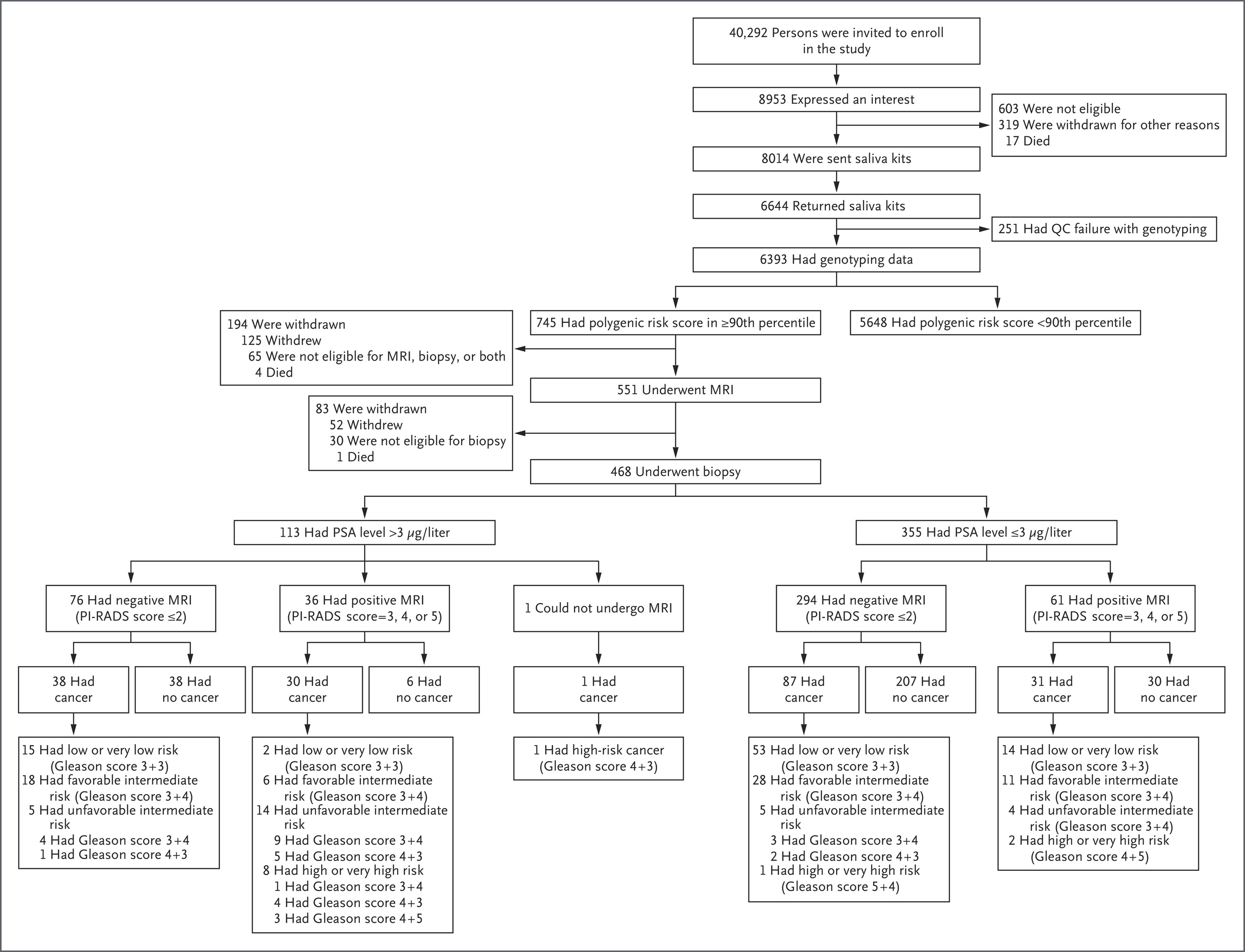
From March to July 2019, over 40,000 men were invited to participate in the BARCODE1 study, with 8,953 (22.2%) expressing interest. Among the 6,393 participants who submitted valid DNA samples, 745 (11.7%) were found to be in the top 10% of the polygenic risk score distribution. Of these, 468 men (62.8%) underwent multiparametric MRI and transperineal biopsy. Prostate cancer was diagnosed in 187 participants (40.0%), with a median age of 64 years at diagnosis.
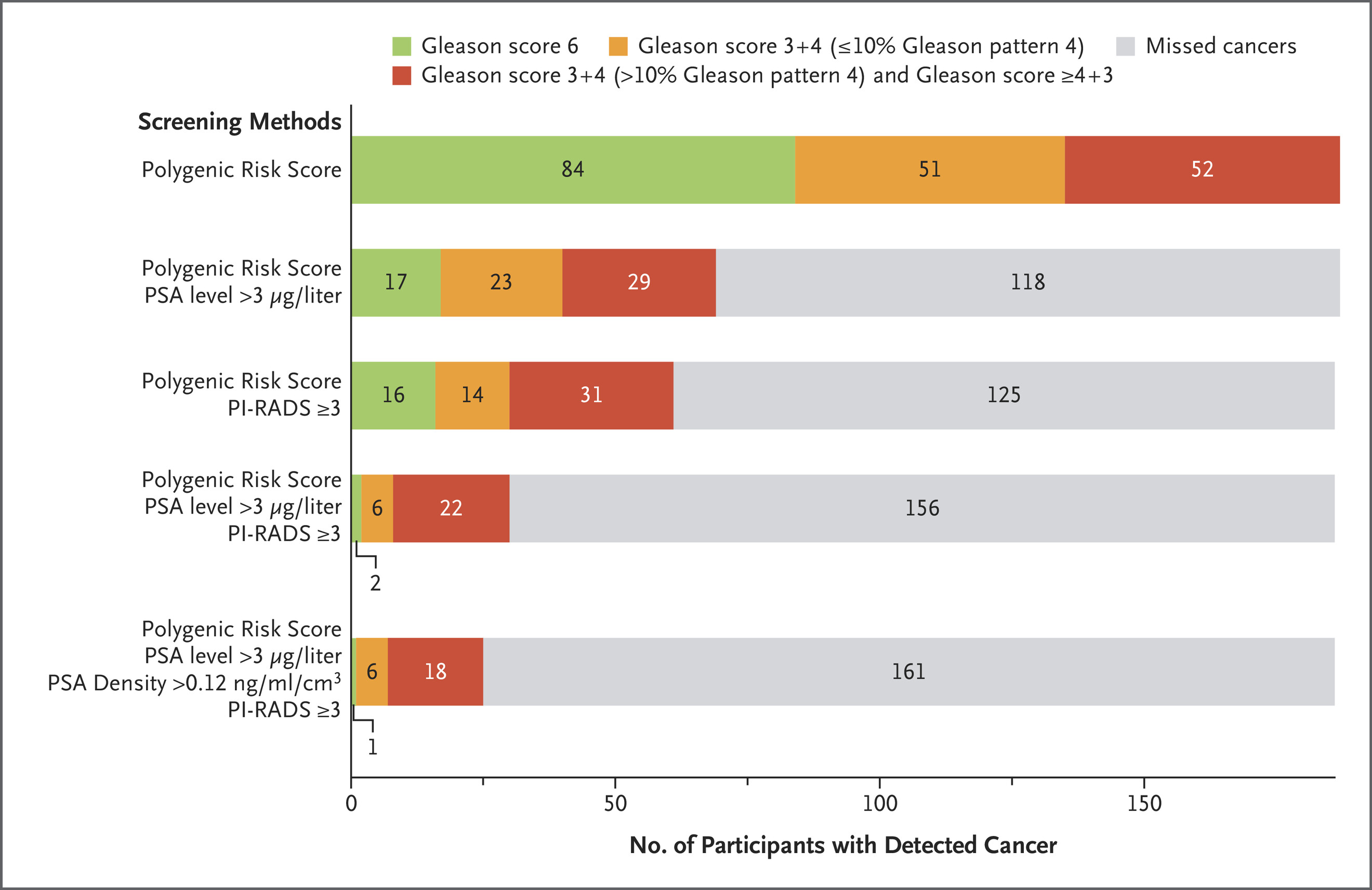
Over half of these cancers (55.1%) were clinically significant (Gleason score ≥7), and 21.4% were classified as unfavorable intermediate, high, or very high risk, necessitating radical treatment. Importantly, 71.8% of clinically significant cancers in this high-PRS group would have been missed using the UK’s standard diagnostic pathway based solely on elevated PSA and MRI findings.
Additionally, 63.1% of all diagnosed cancers had PSA values ≤3.0 μg/L, and 125 cancers were found in men with negative MRI scans, emphasizing the limitations of conventional screening tools. These findings demonstrate that polygenic risk score–based stratification can identify clinically relevant prostate cancer in men who would otherwise not meet biopsy criteria, thereby enhancing detection without increasing unnecessary procedures.
- 6,393 participants had polygenic risk scores calculated; 745 (11.7%) were in the ≥90th percentile
- 468 of the high-PRS participants underwent MRI and biopsy
- Prostate cancer detected in 187 of 468 (40.0%)
- Clinically significant cancers (Gleason ≥7) in 103 of 187 (55.1%)
- 40 of 187 (21.4%) had high-risk cancers requiring radical treatment
- 118 of 187 (63.1%) diagnosed cancers had PSA ≤3.0 μg/L
- 125 cancers were detected in participants with negative MRI (PI-RADS ≤2)
- Only 30 of 187 cancers (16.0%) would have been referred for biopsy under UK standard criteria
- 17 of 40 high-risk cancers (42.5%) would have been missed with standard screening
- Positive predictive value of PSA >3.0 μg/L: 61.1%
- Positive predictive value of MRI (PI-RADS ≥3): 62.9%
- AUC for model combining age, family history, PSA, and MRI: 0.78 for clinically significant cancer
- Estimated overdiagnosis in PRS-based screening: 20.8%, comparable to PSA-based methods
- Adverse events were minimal: 1 case of sepsis (0.2%) and 2 cases of UTI (0.4%) post-biopsy
Key Findings
The BARCODE1 study demonstrates that using a polygenic risk score to target men in the top decile of genetic risk improves prostate cancer detection. This approach detects more clinically significant cancers and avoids many unnecessary diagnoses and biopsies. PRS-based screening may offer a cost-effective, one-time tool to personalize prostate cancer screening. However, further research is needed to validate findings in non-European populations, assess long-term outcomes, and integrate PRS into national screening programs.
- 40% of men with PRS ≥90th percentile who underwent MRI and biopsy were diagnosed with prostate cancer.
- 55.1% of those diagnosed had clinically significant disease.
- PRS-based screening identified high-risk cancers that would have been missed by PSA + MRI alone.
- PSA alone missed 63.1% of cancers.
- MRI missed 125 cancers (including 57 with Gleason ≥7).
- PRS improved patient selection and reduced unnecessary biopsies and overdiagnosis.
Key Takeaway Messages
- PRS enhances the accuracy of prostate cancer screening beyond PSA and MRI alone.
- A one-time genetic test enables lifelong risk stratification.
- Personalized screening strategies using PRS can reduce harms of overdiagnosis while catching more clinically significant cancers.
- PRS is especially valuable in individuals with normal PSA or negative MRI but genetically high risk.
You can read the full article here


