Thomas Köhnke, Instructor at Institute for Stem Cell Biology and Regenerative Medicine, shared a post on X:
“Excited to share our work on ASXL1 mutations in clonal hematopoiesis and myeloid malignancies, out in Blood Cancer Discovery.
Blood Cancer Discovery
Mutations in ASXL1 are found both in clonal hematopoiesis as well as a variety of myeloid malignancies and over the past decade, both loss-of-function as well as gain-of-function mechanisms of clonal expansion have been proposed.
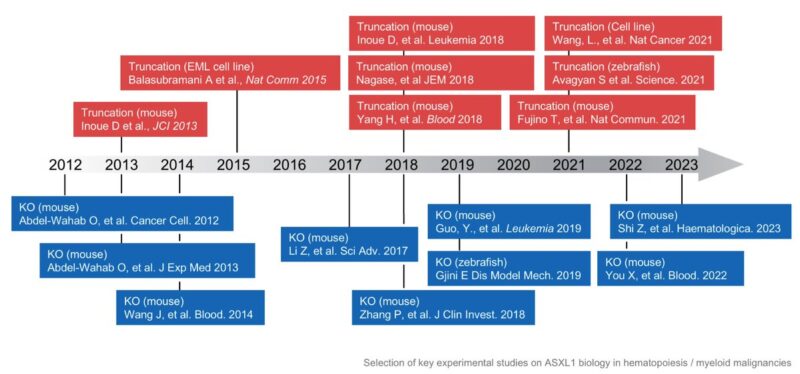
Seminal experimental studies in the early 2010s, including work by Abdel-Wahab Lab (in Ross Levine‘s lab) studied deletion of ASXL1, whereas others, including Daichi Inoue
(in Toshio Kitamura’s lab) investigated truncating mutations in murine hematopoiesis.
Since then, several groups have leveraged either deletion or truncation-based models to investigate the role of ASXL1 mutations, however a definitive consensus has not emerged and both continue to be utilized.
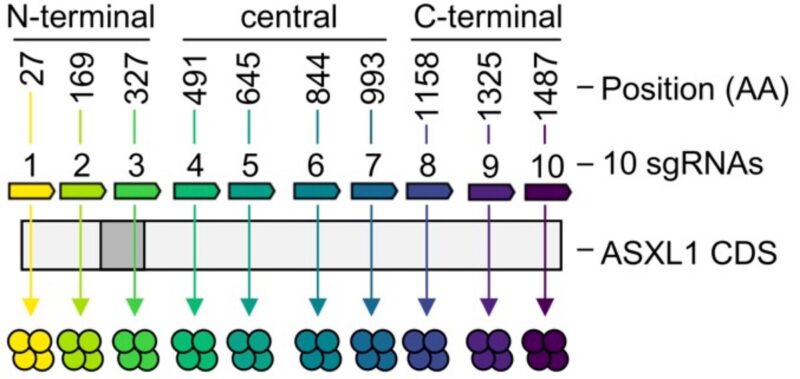
Here, we hope to contribute some clarifying studies using human primary cells. Using CRISPR/Cas9 in human HSPCs, we interrogated the entire ASXL1 coding sequence and confirmed that only frame-shift lesions in the center of the coding sequence enhance stemness and myeloid bias.
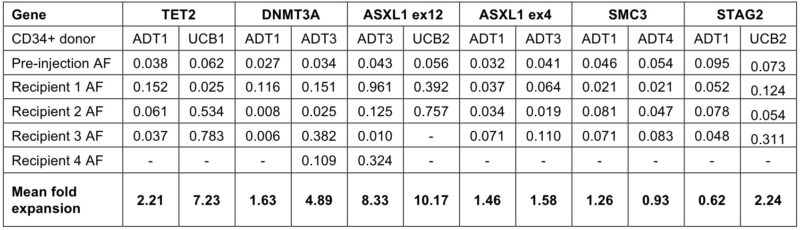
Further, we compared deletion versus truncation of ASXL1 directly, showing that only truncating ASXL1 mutations are associated with defective differentiation along erythroid- and megakaryocytic lineages, and only truncating ASXL1 mutations show a competitive advantage in vivo.
This more definitively confirms some of the few direct, prospective comparisons such as by Tothova Lab
(in Ben Ebert’s lab, published in Cell Stem Cell 2017) – where there is greater expansion in vivo for exon 12 mutations (data is somewhat hidden in the supplemental table S1B).
Molecularly, we confirmed increased activity of the ASXL1-BAP1 (PR-DUB) complex when ASXL1 is truncated, leading to reduced global levels of the repressive H2AK119Ub chromatin mark in human HSPCs.
Using CUT&RUN, we profiled the distribution of H2AK119Ub across the genome and found that while H2AK119Ub is globally reduced, this is not uniform. Rather, low-occupancy regions are being further depleted, whereas high-occupancy regions – intriguingly – having increased H2AK119Ub.
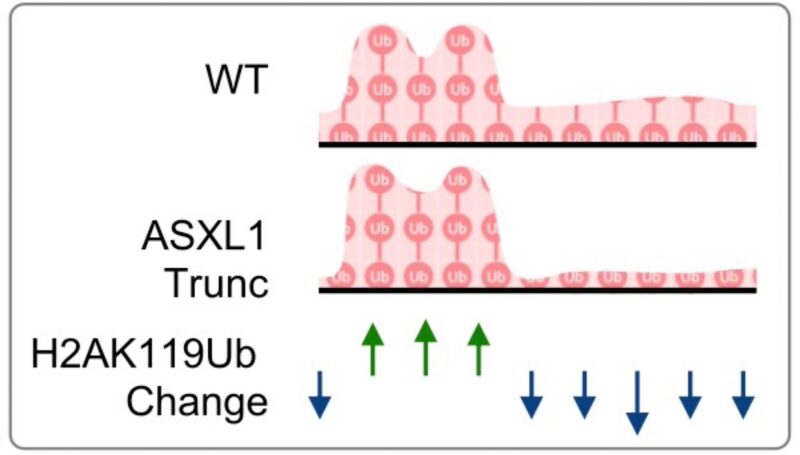
This finding fits with our more general understanding of PR-DUBs role – which is thought to limit the genome-wide accumulation of H2AK119Ub, and thus being central to Polycomb complex occupancy at Polycomb target genes.
We further validate this pattern of H2AK119Ub redistribution both in isogenic cell lines as well as primary human AML samples and show that when PR-DUB members are deleted, this pattern is inverted.
Finally, we interrogate whether this balance of H2AK119Ub deposition (by PRC1) and removal (by PR-DUB) is influenced by the expression levels of truncated ASXL1 and find a “Goldilocks” zone of expression.
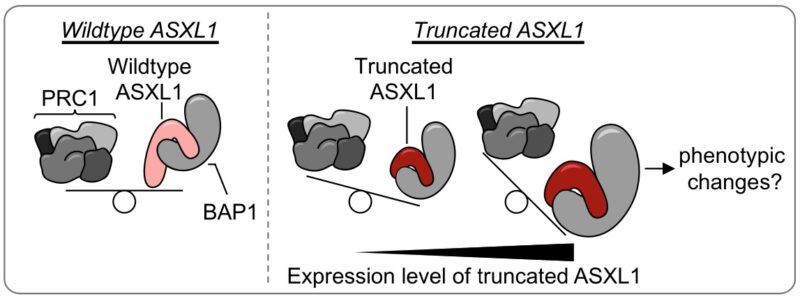
This feature might explain why mutations in ASXL1 are almost exclusively heterozygous in clonal hematopoiesis and myeloid disease.
Finally, we interrogated whether this balance of H2AK119Ub deposition and removal can be exploited therapeutically, and indeed, cells with truncated ASXL1 are more vulnerable to PRC1 inhibition; both in our primary HSPCs as well as patient samples.
Hopefully our data can lead to further exploration of the role of ASXL1 in disease initiation and maintenance. Thank you to Ravi, my co-authors in the Majeti Lab, and funding from Leukemia & Lymphoma Society Research and the EvansMDS foundation for allowing us to work on this project for the past years.”
Source: Thomas Köhnke/X
