
New Paper Alert! Proteogenomic Analysis Identified New Cancer Drug Targets Across 10 Cancer Types
Proteogenomic Analysis Identified New Cancer Drug Targets Across 10 Cancer Types .
Authors: Sara R. Savage, Xinpei Yi, Jonathan T. Lei, Bo Wen, Hongwei Zhao, Yuxing Liao, Eric J. Jaehnig, Lauren K. Somes, Paul W. Shafer, Tobie D. Lee, Zile Fu, Yongchao Dou, Zhiao Shi, Daming Gao, Valentina Hoyos, Qiang Gao and Bing Zhang.
Published in Cell, on June 24, 2024
Introduction:
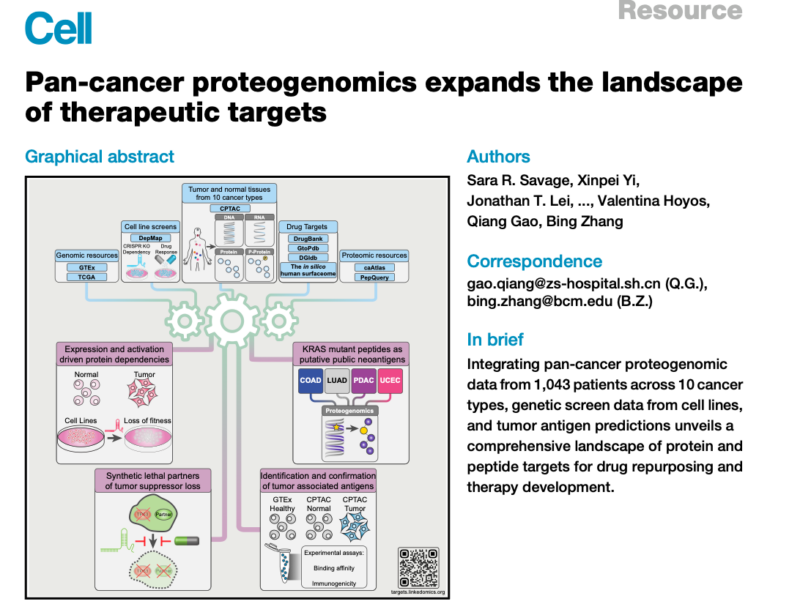
This novel study has expanded our understanding of potential therapeutic targets for cancer treatment. Researchers integrated proteogenomic data from over 1,000 patients across 10 cancer types with external datasets to identify and prioritize new drug targets. This comprehensive analysis provides valuable insights into existing cancer drug targets and reveals promising new avenues for therapy development.
Design/Methods:
The study analyzed harmonized Clinical Proteomic Tumor Analysis Consortium (CPTAC) proteogenomics data from 1,043 tumor samples and 524 normal tissue samples across 10 cancer types. The researchers integrated multiple omics data types, including mutation, copy-number variation, methylation, transcript, protein, and phosphosite abundance data. They also incorporated data from external sources such as the Cancer Dependency Map (DepMap) and drug response data from the PRISM drug repurposing resource.
The team employed various computational approaches, including differential abundance analysis, kinase-substrate enrichment analysis, and neoantigen prediction. They also used experimental validation techniques, such as shRNA knockdown, drug response assays, and immunogenicity testing.
What We Learned:
- Proteomic Landscape of Drug Targets: The study quantified 2,863 druggable proteins, revealing a wide range of protein abundances across cancer types. Interestingly, druggable genes showed higher median mRNA-protein correlations compared to other genes, but over half still had correlations below 0.6.
- Targetable Dependencies: Researchers identified 51 proteins overexpressed in at least 5 cancer types that also showed dependency in cell line experiments. Integration with genomic data revealed proteins whose overexpression was associated with mutations, hypomethylation, or copy-number amplification.
- Protein Hyperactivation: Analysis of phosphosite data identified 31 protein hyperactivation events occurring in two or more cancer types. The study also inferred altered kinase activity, with 19 kinases showing increased activity across multiple cohorts.
- Evaluation of Predictions: The team’s method for predicting druggable dependencies achieved a 39% success rate in identifying effective drug responses, a 2.6-fold increase over random selection. This approach showed 83% specificity and 76% accuracy in predicting drug responses.
- Tumor Suppressor Gene Loss: The study identified protein dependencies associated with the loss of tumor suppressor genes, revealing potential synthetic lethal therapeutic strategies. For example, TOP2A overexpression was linked to TP53 loss in uterine cancer.
- Neoantigen Identification: Study results predicted 2,315 putative neoantigens associated with 846 somatic mutations. Notably, 5 KRAS mutant peptides were predicted to yield neoepitopes in 44 patients across 4 cancer types.
- Tumor-Associated Antigens: The study identified 140 proteins with highly restricted expression in normal tissues but abnormal expression in tumors. Experimental validation of selected peptides revealed 22 with both strong binding affinity and immunogenicity.
Key Highlights:
- Integration of proteomics data with other omics types provided insights into protein targets not evident from genomics alone.
- Pan-cancer analysis identified common therapeutic targets relevant to multiple cancer types.
- The study’s predictive approach for druggable dependencies showed high specificity and accuracy.
- Identification of KRAS mutant peptides as potential public neoantigens across multiple cancer types.
- Experimental validation of tumor-associated antigen peptides revealed promising immunotherapy targets.
Key Takeaway Messages:
- Proteogenomic integration offers a powerful approach for identifying and prioritizing cancer drug targets.
- Pan-cancer analysis can reveal common therapeutic targets that may be overlooked in single cancer type studies.
- Combining tumor profiling data with cell line perturbation experiments enhances the identification of clinically relevant targets.
- The study provides a comprehensive resource of protein and peptide targets covering various therapeutic modalities.
- The findings pave the way for drug repurposing and new therapy development across multiple cancer types.
This extensive proteogenomic analysis has significantly expanded the landscape of potential cancer therapeutic targets. As we move towards more personalized and effective cancer treatments, such comprehensive analyses will play a crucial role in identifying the most promising targets for future clinical investigation.
Summary by Amalya Sargsyan, MD
Proteogenomic Analysis Identified New Cancer Drug Targets Across 10 Cancer Types
