
Itai Yanai: ‘Resistance continuum’ of resistant phenotypes of cancer cells
Itai Yanai, Professor at the Department of Biochemistry and Molecular Pharmacology at NYU, shared on X:
“Published today! We found that cancer cells adapting to drug treatments don’t simply switch from a sensitive to resistant state; instead there’s a ‘resistance continuum’ of resistant phenotypes with epigenetically reprogrammed states.
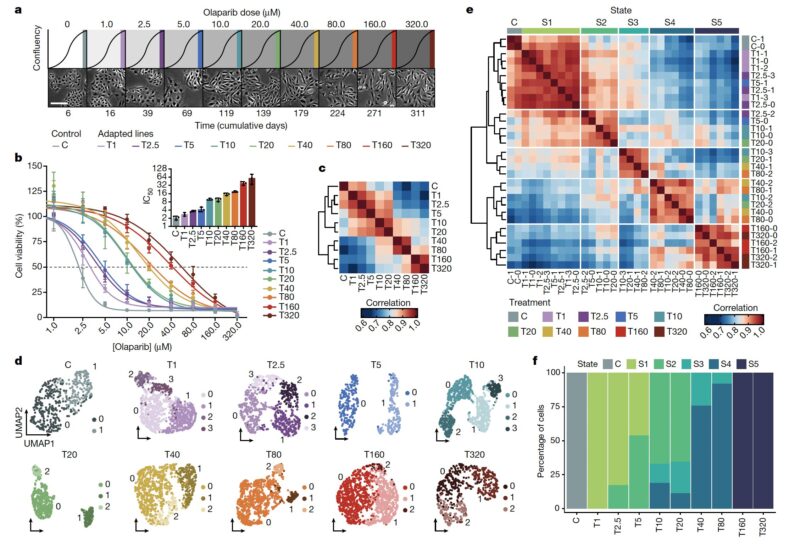
We found that cellular adaptation occurs by a series of cell state transitions, each with distinct gene expression programs and leading to increased resistance. This follows on the pioneering work of Sydney Shaffer, Arjun Raj, Andriy Marusyk, Marine Lab and others.
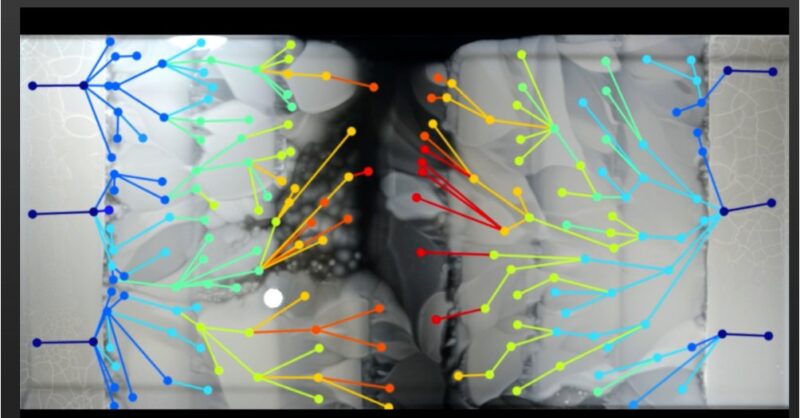
Our inspiration was the famous ‘MEGA-plate experiment’ by Michael Baym, Tami Lieberman, Roy Kishony et al, in which bacteria were grown on a set of antibiotic concentrations. Exposure to low concentrations facilitated resistance to higher concentrations.
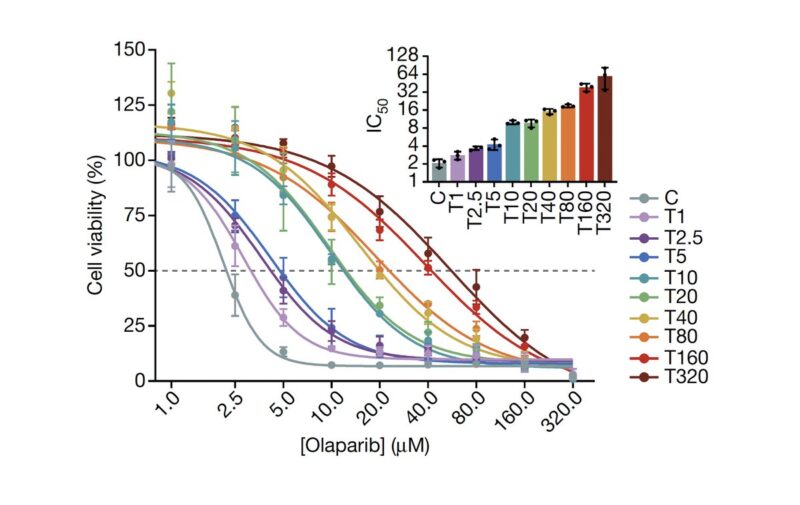
We imagined that an analogous process may be happening in cancer cells, and so we performed a dose-escalation experiment on ovarian cancer cells treated with a PARP inhibitor. Over the course of 12 months we generated a panel of 10 adapted populations.
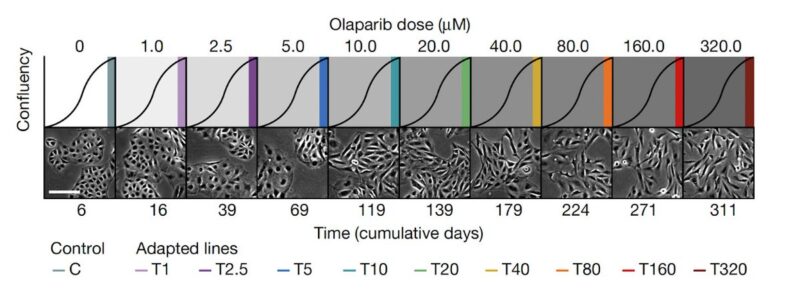
Similarly to the MEGA-plate experiment, we found that previous drug exposure with lower doses facilitated the emergence of resistant populations to progressively higher doses. Thus, incremental adaptive responses unfolded along a continuum, and not a transition to full resistance.
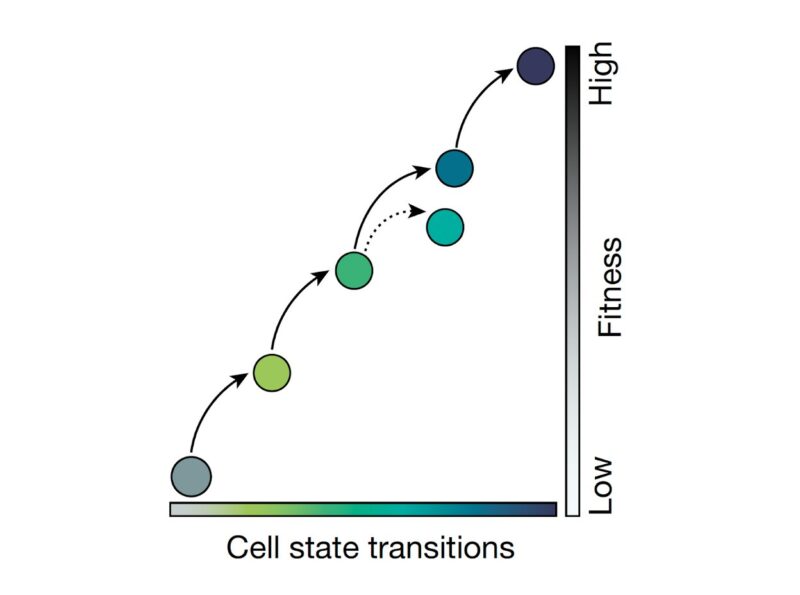
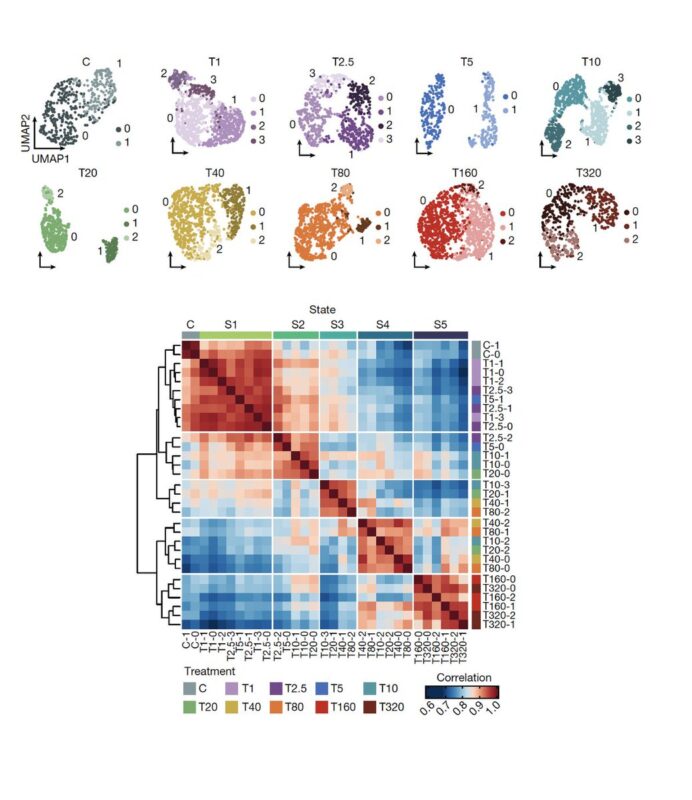
Applying single-cell RNA-Seq, we found that as cancer cells become increasingly drug resistant, their gene expression programs evolved accordingly by the emergence of new transcriptional states, and multi-step state transitions among them.
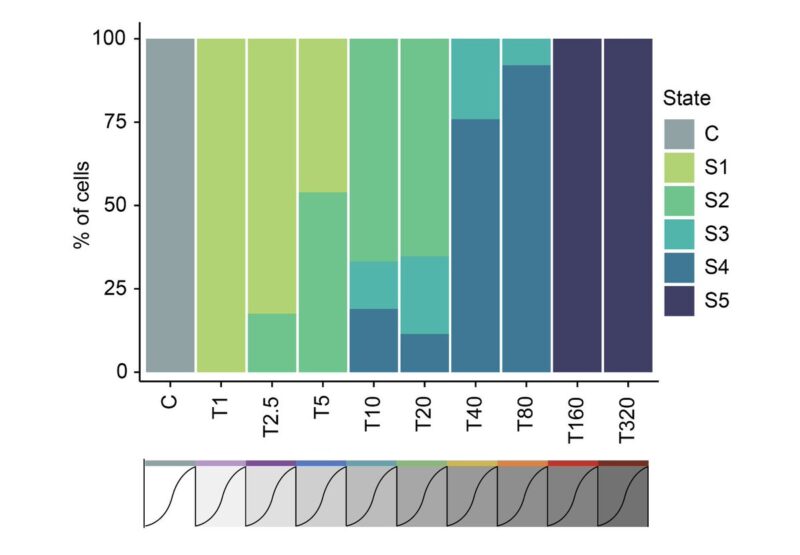
Cellular adaptation occurs along a continuum driven by the emergence and selection of cellular states. You can see this better when looking at the frequency of cells in each of the five states across the adaptive lines.
Using a gene module detection approach, we detected the gene expression modules that underlie the states. These gene modules each comprise distinct biological functions that may contribute to the survival of the drug resistant cells.
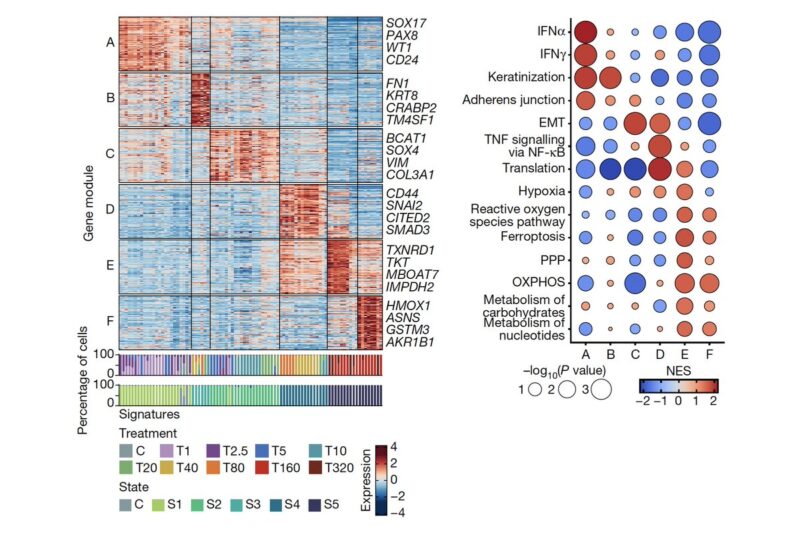
We also found that epithelial-to-mesenchymal transition (EMT) or stemness programs—often considered a proxy for phenotypic plasticity—enable adaptation, but are not a full resistance mechanism. We particularly thank Andriy Marusyk
& Alicia Bjornberg for working with us on this!
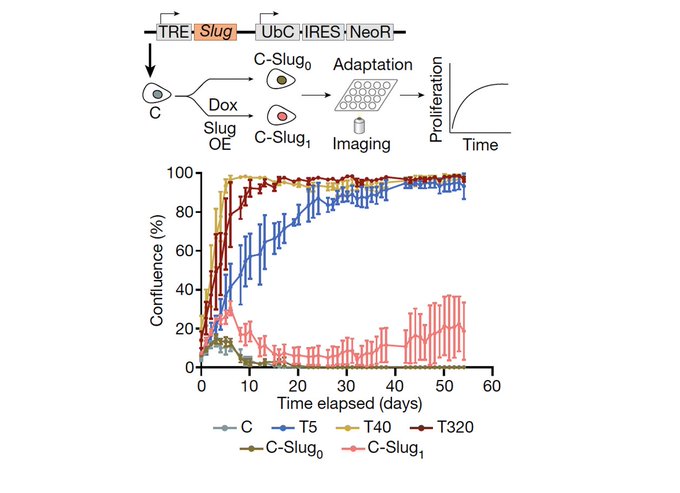
Is this genetic? We found that copy number alterations are indeed associated with the states, but also that these are reinforced by epigenetically regulated stress response TFs (AP1, NRF2, ATF4) & the loss of cell identity in terms of closing sites associated with lineage markers.
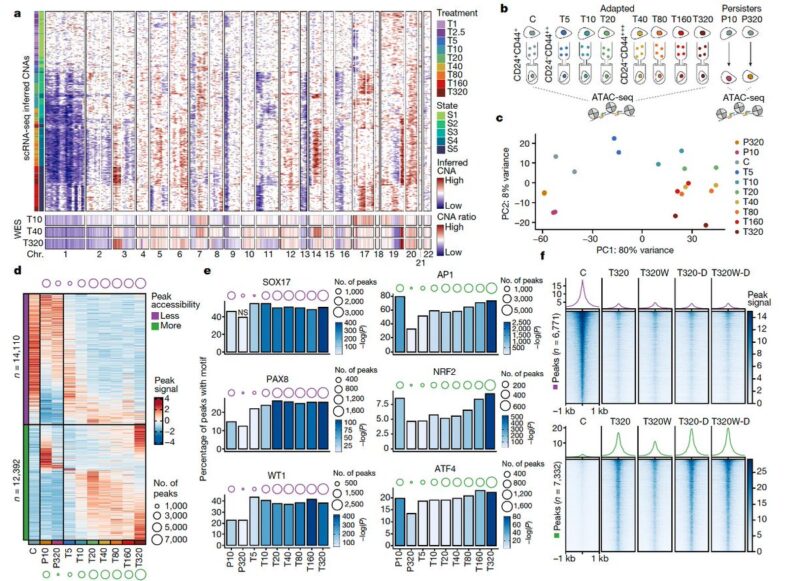
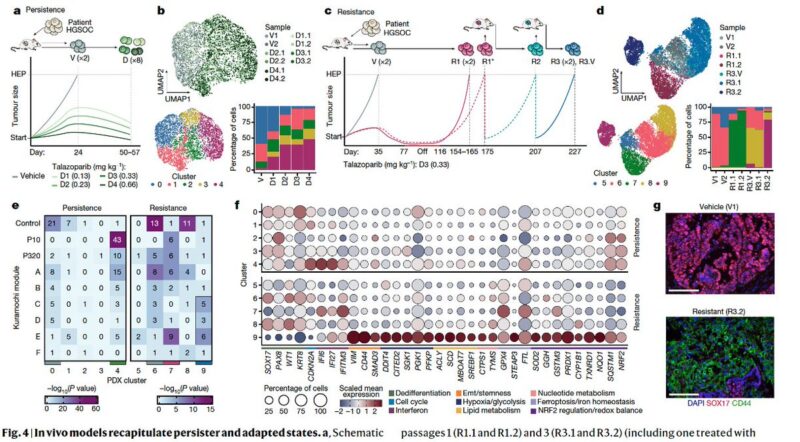
To study this in vivo, we generated two mouse models for shorter (persistence) and longer term response (resistance). We found that the short-term treatment elicited a partial reprogramming while the resistance model revealed convergence with the in vitro adapted states.
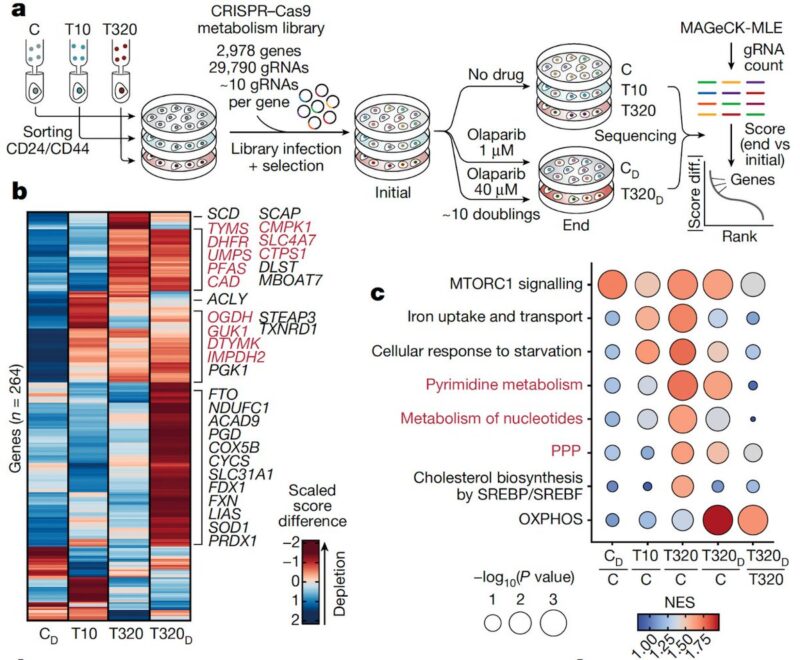
We also performed a negative selection CRISPR screen using a metabolic gene knockout library to test whether the genes that are expressed in the resistant states are also required. In line with the adaptive modules, we found an increased dependency on nucleotide metabolism.
These dynamics of cellular adaptation may have clinical implications in the sense that limited penetration of many drugs into solid tumors generates real-life drug gradients, thus possibly priming cells for adaptation.
Our study also highlights the need for careful dose optimization, treatment intervals and longitudinal monitoring aiming to prevent drug-induced adaptation.
This paper is a testament to the amazing work of Gustavo S. França who spearheaded this entire project! Gustavo is also on the job market now by the way.

It was also a great collaboration with Tim Lionnet‘s lab, in particular with Ben King, and with the labs of Papagiannakopoulos Lab and Andriy Marusyk, with the dedicated work of Jozef Bossowski and Alicia Bjornberg.
It really takes a village to do a project like this. Thank you for the amazing contributions of Maayan Baron, Maayan Pour, Anjali Rao, Selim Misirlioglu, Dalia Barkley, Igor Dolgalev, Kwan Ho Tang, Marta Chiodin, Gal Avital, Felicia Kuperwaser, Ayushi Patel, Debbie Liberman, Douglas Levine.”
Read the study.
Source: Itai Yanai/X
